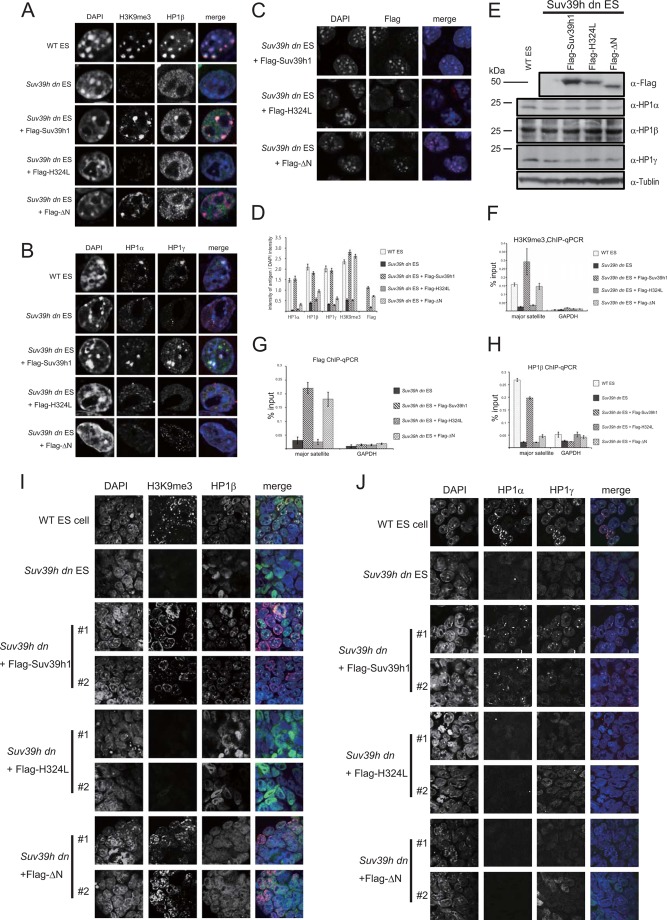
FIGURE 2.
Suv39h1ΔN could accumulate and deposit H3K9me3 at the pericentromere, but HP1 was generally absent from the pericentric region. Immunohistochemical staining analysis for H3K9me3 (red) and HP1β (green) (A), HP1α (red) and HP1γ (green) (B), or FLAG-tagged Suv39h1 WT, H324L, or ΔN (red) (C) was conducted with WT or Suv39h dn ES cells or Suv39h dn ES cells complemented with either FLAG-tagged WT Suv39h1, H324L, or ΔN. DNA was counterstained with DAPI (blue). D, DAPI intensity and the intensity of the antigen at the same region were quantified using ImageJ based on the fluorescence imaging data obtained for A–C. The intensities for 30 cells were quantified. The value of each antigen intensity was expressed as a ratio between the DAPI intensity and the intensity of the antigen being measured. E, Western blot analysis of endogenous HP1 subtypes in the indicated cell lines. F–H, chromatin was isolated from the indicated cell lines. ChIP-quantitative PCR analysis was conducted using anti-H3K9me3 (F), anti-FLAG (G), anti-HP1β (H), and primers specific for major satellite repeats and the Gapdh gene. I and J, immunohistochemical staining analysis for the two independent cell lines indicated at low magnification. A combination of color and antigen was same as that observed in A and B.