Figure 3.
Transcriptional Profiles Confirm Accelerated Differentiation in WNT3A/BMP4 EBs
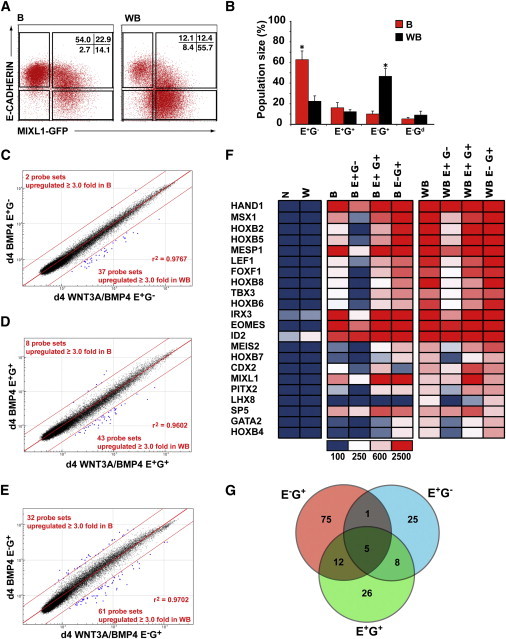
(A) Flow cytometry plots showing the position of gates used for sorting experiments based on E-CADHERIN and MIXL1-GFP expression in d4 EBs differentiated in APEL medium supplemented with BMP4 (B) and WNT3A/BMP4 (WB). The percentage of cells falling into each quadrant is shown.
(B) Histogram showing the mean percentage of cells in E-CADHERIN- (E) and MIXL1-GFP- (G) expressing populations. Data represent the mean ± SEM from four independent experiments. B and WB groups were compared using Student’s t test. *p < 0.05.
(C–E) Comparison of transcriptional profiles of sorted cell populations shown in (A) and (B) derived from BMP4- and WNT3A/BMP4-stimulated cultures. Enhanced dots outside the parallel red lines indicate probe sets differing by ≥3-fold from the mean.
(F) Heatmap showing the expression of transcription factors upregulated in d4 WNT3A/BMP4 E−G+ cells. These primitive streak or posterior mesodermal genes were frequently expressed at a similar level in the corresponding BMP4 sorted fraction. The scale for mean signal intensity in arbitrary units is shown.
(G) Venn diagram illustrating the overlap between the differentially expressed probe sets in the sorted populations identified in (C)–(E). The genes recognized by the E−G+ and E+G+ probe sets and their level of expression are listed in Tables S3 and S4.
See also Figures S4 and S5.
