Figure 2.
Genome-wide Analysis of Imprinted DMRs in PSCs
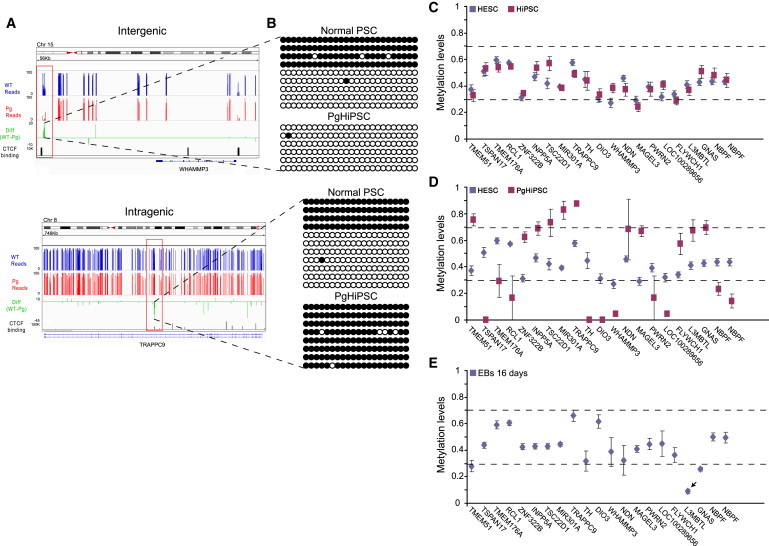
(A) Regional view of two representative new DMR, TRAPPC9 and WHAMMP3. Average methylation values for wild-type HESCs (blue) and PgHiPSCs (red) of all CpG calls. Green track indicates the difference between hemimethylated (AMC between 0.3 and 0.7) wild-type PSCs and PgHiPSCs CpGs. Shown are significant putative CTCF binding sites in H1 ES cells from the ENCODE project (depicted in black rectangles, p value < 1 × 10−5) and CpG islands (UCSC) in dark-green rectangles. See also Figure S3.
(B) Bisulfite sequencing validation of pDMR (WHAMMP3, upper panel) and mDMR (TRAPPC9, lower panel) was conducted on normal independent PSC not included in the original analysis and PgHiPSCs lines.
(C–E) Methylation values (y axis) ± SE in various cell types across the different imprinted DMRs (x axis); small arrows pointing to DMRs that are perturbed in the different cell types. (C) Comparison between HESCs and HiPSCs demonstrate the striking similarities between the two cell types. (D) Comparison between HESCs and the PgHiPSCs showing the differences between the two cell types in methylation values, as the PgHiPSCs are either hypermethylated (AMC >0.7) or hypomethylated (AMC <0.3) in comparison to hemimethylation state (AMC between 0.3 and 0.7) of the biparental cells. (E) Analysis of methylation in 16-day-old EBs differentiated from HESCs, demonstrating that the newly identified imprinted DMRs are highly stable following in vitro differentiation. Notable exception is the DMR in the L3MBTL locus, which is consistently hypomethylated in the differentiated cells.
