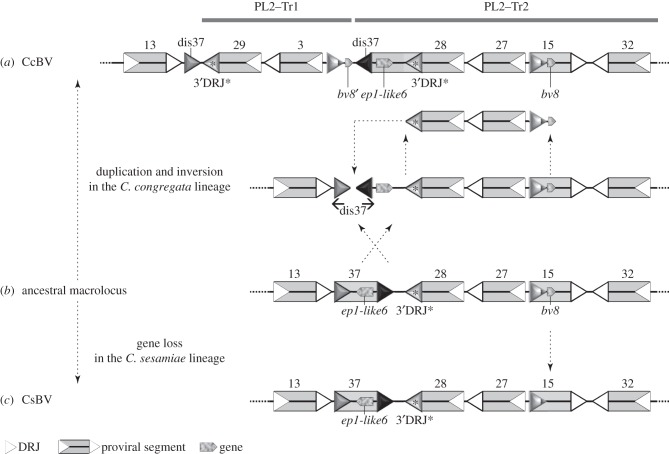
Figure 7.
A proposed parsimonious scenario for the complex rearrangement that may have produced PL2-Tr1 based on the analysis of duplications among Cotesia spp. (a) Cotesia congregata and (c) C. sesamiae macrolocus sequences were used to infer the putative organization of this region in their common ancestor (b). In the lineage leading to C. sesamiae, the bv8 gene was lost (or this gene was acquired specifically in the C. congregata lineage). In the lineage leading to C. congregata, a complex rearrangement occurred resulting in inversion and duplication of proviral segment sequences. Inversion: the segment S37 was inverted and its ep1-like6 gene and regular 3′DRJ (black triangle) were incorporated into an enlarged S28. The regular 3′DRJ became that of S28, replacing the former S28 DRJ readily identified by its particular sequence (3′DRJ*, grey triangle). Duplication: the region encompassing S28, S27 and a part of S15 was duplicated and inserted within S37 that was dismantled (dis37). It should be noted that inversion, duplication and dismantlement might have been produced by a single complex rearrangement caused by errors during replication (fork stalling and template switching model). DRJs are indicated by white triangles to delimit the segments.