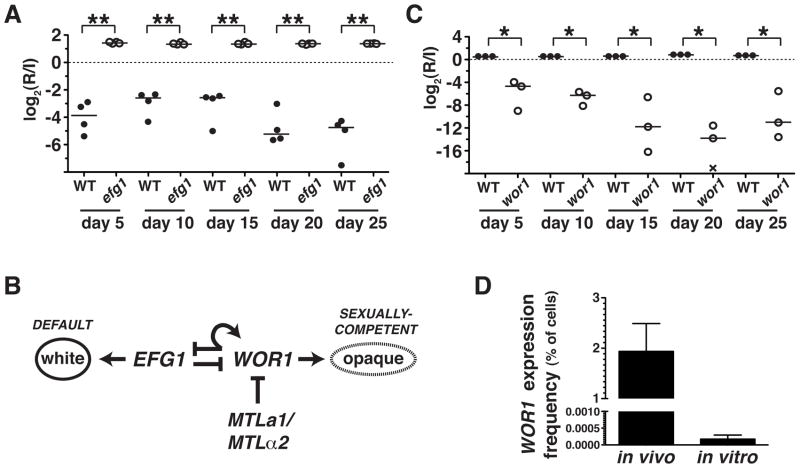
Figure 1. EFG1 inhibits and WOR1 promotes C. albicans fitness in the commensal milieu.
A) Competition experiment between wild-type (WT, SN250) and efg1ΔΔ mutant (SN1011) strains in 4 mice. Relative abundance was determined by qPCR, using strain-specific primers. Log2(R/I) represents the log ratio of the abundance of each strain after recovery from the feces of a given animal (R) compared to the level in the infecting inoculum (I). * indicates p<0.001 by the t-test (two-tailed, unpaired samples). Similar results were obtained when the efg1ΔΔ mutant was competed in a pool of 48 strains, as described in the text (Pande and Noble, unpublished). B) Transcriptional regulation of the white-to-opaque switch. C) Competition experiment between WT (SN250) and wor1ΔΔ (SN881) in 3 mice, as above. * p<0.05. Similar results were obtained in tests of two additional wor1ΔΔ strains, shown in Supplementary Figure 1, A–B. D) Comparison of WOR1 expression in MTLa/α cells grown in the commensal model or in vitro. The WOR1promoter-FLP strain (SN1020) was propagated for 3 days in the murine model or for 8 generations in liquid culture medium, followed by 5-FOA selection and PCR of 5-FOAR colonies to verify Flp-mediated deletion URA3. Mean percentages of cells meeting these criteria are plotted on the y-axis. Error bars reflect the standard deviation among 6 (in vivo) or 4 (in vitro) biological replicates.