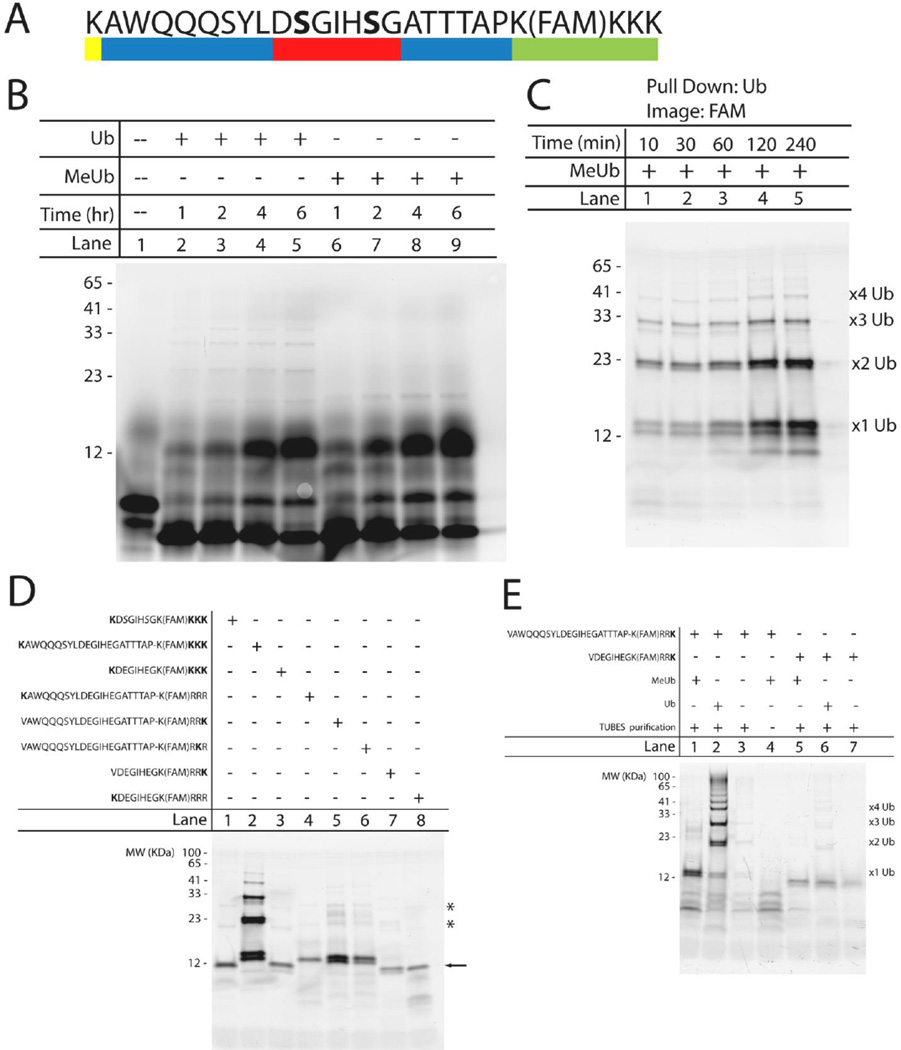
Figure 4. Development of a novel UPS substrate based on the β-Catenin degron.
(A) Sequence of the degron-based substrate containing four essential components: the β-Catenin degron (red), an ubiquitination site lysine (yellow), amino acid spacing sequences to prevent steric hindrance (blue), and a fluorescein tag for detection (green). The bold serines are phosphorylated. (B) Time-dependent ubiquitination of the degron-based substrate using either ubiquitin (lanes 2–5) or methylated ubiquitin (lanes 6–9). Lane 1 contains unmodified substrate (∼3.1 kDa). All bands were detected using a Typhon Imager (GE Healthcare Life Sciences) to visualize the 5,6 carboxyfluorescein tag. Numbers to the left of the gel correspond to the apparent molecular weight (in kDa) determined by the known Benchmark fluorescent protein standard (Invitrogen). (C) Verification of substrate ubiquitination using TUBES to selectively pull down ubiquitinated substrate at the indicated times. Assay samples were incubated with methylated ubiquitin. Suspected mono-, di-, tri-, and tetra-ubiquitinated substrates are labeled accordingly. (D) Iterative approach to identify the importance of proximity and location of ubiquitination site lysine on substrate ubiquitination. All samples were incubated in the in vitro ubiquitination assay for 2 hours with methylated ubiquitin prior to pull down with TUBES. (E) Two degron-based substrates, containing a single C-terminal ubiquitination site lysine, were incubated in the in vitro ubiquitination assay, along with TUBES pull down, under the following conditions: methylated ubiquitin (lane 1,5), ubiquitin (lane 2,6), no exogenous MeUb or Ub (lane 3,7) or control-agarose beads instead of the agarose-TUBES (lane 4). Mono-, di-, tri, and tetra-ubiquitinated species are labeled accordingly.