Fig. 8.
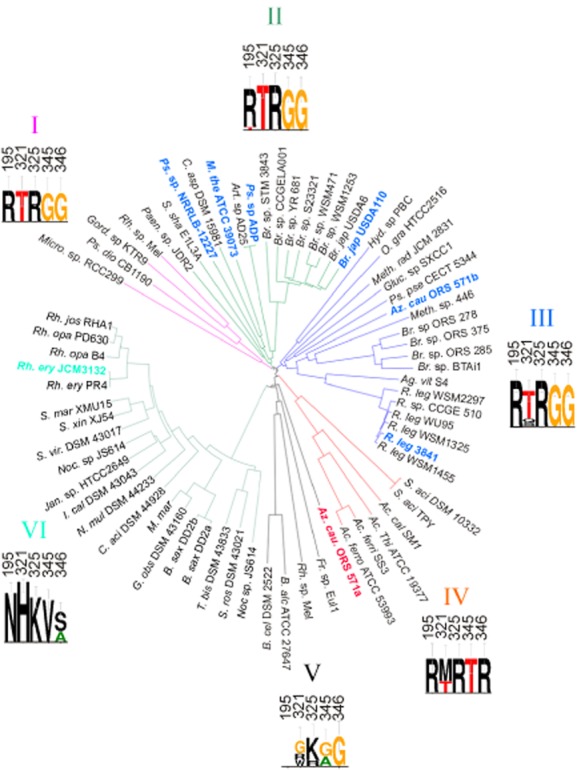
Phylogeny of the AtzD/Bar family. The sequences for the AtzD/Bar homologues in the six groups were sourced from the following organisms (listed in order, clockwise): Group I (purple); Micromonas sp. RC299, Pseudomonas dioxanivorans CB1190, Gordonia sp. KTR8, Rhodococcus sp. Mel. Group II (green); Paenibacillus sp. JDR2, Salinisphaera shabanensis E1L3A, Pseudomonas sp. NRRLB-12227, Clostridium asparagiforme DSM 15981, Moorellia thermoacetica ATCC 39073, Arthrobacter sp. AD25, Pseudomonas sp. ADP, Bradyrhizobium sp. STM3843, Bradyrhizobium sp. CCGELA001, Bradyrhizobium sp. YR681, Bradyrhizobium sp. S23321, Bradyrhizobium sp. WSM471, Bradyrhizobium sp. WSM1253 Bradyrhizobium japonicum USDA6, Bradyrhizobium japonicum USDA110. Group III (blue); Hydrogenophaga sp. PBC, Oceanicoia granulosus HTCC2516, Methylobacterium radiotolerans JCM 2831, Pseudomonas pseudoalcaligenes CECT 5344, Azorhizobium caulinodans ORS 571, Methylobacterium sp. 446, Bradyrhizobium sp. ORS 278, Bradyrhizobium sp. ORS 375, Bradyrhizobium sp. ORS 285, Bradyrhizobium sp. BTAi1, Agrobacterium vitis S4, Rhizobium leguminosarum bv. trifolii WSM2297, Rhizobium sp. CCGE 510, Rhizobium leguminosarum bv. trifolii WU95, Rhizobium leguminosarum bv. trifolii WSM1325, Rhizobium leguminosarum bv. viciae 3841, Rhizobium leguminosarum bv. viciae WSM1455. Group IV (red): Sulfobacillus acidophilus DSM 10332, Sulfobacillus acidophilus TPY, Acidithiobacillus caldus SM1, Acidithiobacillus thiooxidans ATCC 19377, Acidithiobacillus ferrivorans SS3, Acidithiobacillus ferrooxidans ATCC 27647. Group V (black): Azorhizobium caulinodans ORS571, Frankia sp. Eul1c, Rhodococcus sp. Mel, Bacillus alcalophilus ATC 27647, Bacillus cellulolytitcus DSM 2552. Group VI (cyan): Nocardioides sp. JS614, Streptosporangium roseum DSM 43021, Thermobispora bispora DSM 43833, Blastococcus saxobsidens DD2, Blastococcus saxobsidens DD2, Geodermatophilus obscures DSM 44928, Modestobacter marinus, Catenulispora acidophia DSM 44928, Nakamurella multipartita DSM 43233, Intrasporangium calvum DSM 43043, Janibacter sp. HTCC2649, Nocardioides sp. JS614, Saccharomonospora viridis DSM 43017, Saccharomonospora xinjiangensis XJ54, Saccharomonospora marina XMU15, Rhodococcus erythropolis PR4, Rhodococcus erythropolis JCM3132, Rhodococcus opacus B4, Rhodococcus opacus PD630, Rhodococcus jostii RHA1. The identities and conservation of amino acids found in each at positions equivalent to AtzD 195, 321, 325, 345 and 346 are shown as logos. The names of bacterial species from which Toblerone fold enzymes have had their catalytic activities characterized are emboldened and coloured to indicate their substrate specificity: blue for cyanuric acid hydrolase, cyan for barbituric acid hydrolase and red for the absence of hydrolytic activity with either cyanuric acid or barbituric acid.
