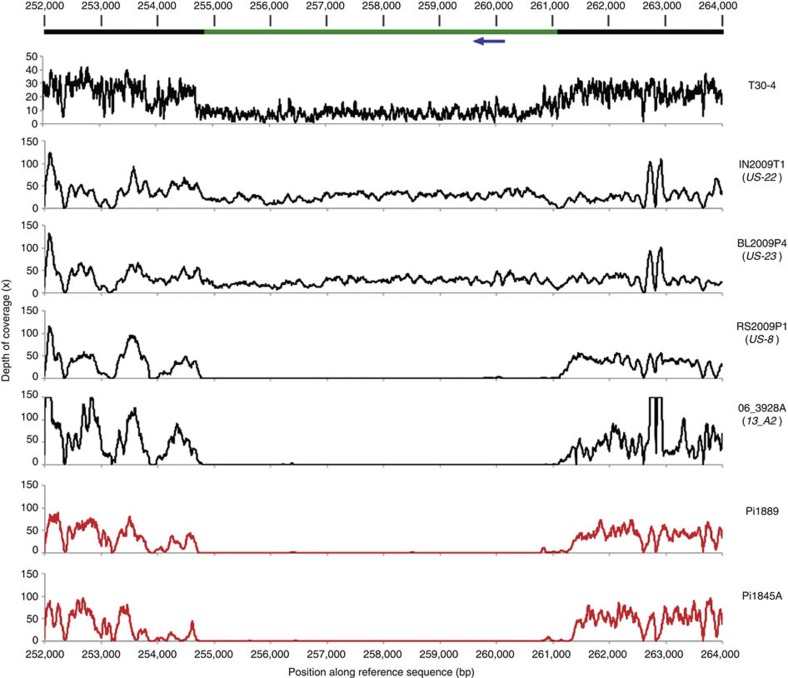
Figure 3. Read depth of coverage around a deleted RXLR effector gene.
The top bar represents the T30-4 strain reference genome assembly sequence for supercontig 66. The green portion of the bar represents a ~6-kbp region deleted in the lower four samples. The blue arrow represents the position of RXLR effector PITG_18215, which is absent from samples with the deletion. Data for the historical samples are plotted in red. Coverage of this region in strain T30-4 is reduced to ~50% of the genome-wide mean. In T30-4, this region is likely either hemizygous or a duplication erroneously absent from the genome assembly. In any case, the sharply defined borders and consistent lack of coverage over the entire deleted region (spanning the length of ~85 successive 70-bp reads) in all samples are conclusive evidence that the region is deleted in four of the resequenced samples.