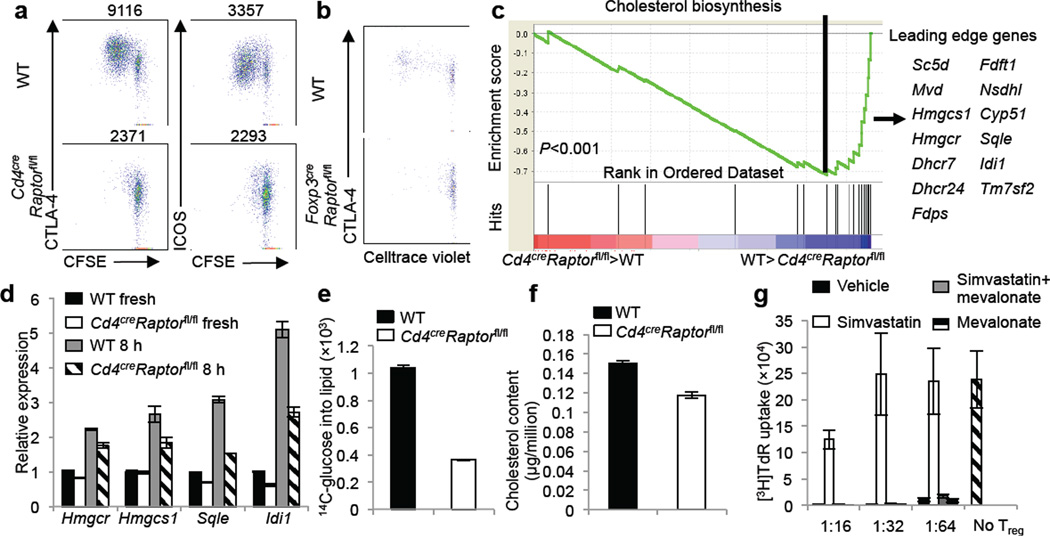
Figure 3.
Raptor coordinates Treg proliferation and effector molecule expression by orchestrating cholesterol/lipid biosynthetic metabolism, especially the mevalonate pathway. a, CFSE-labeled Tregs from wild-type and CD4creRaptorfl/fl mice were stimulated with anti-CD3, anti-CD28 and IL-2 for 3 days, followed by analysis for CFSE dilution and CTLA-4 and ICOS expression (MFI presented above the plots). b, Celltrace violet-labeled Tregs from wild-type and Foxp3creRaptorfl/fl mice were transferred into unmanipulated CD45.1+, and Celltrace violet dilution and CTLA-4 expression were analyzed 7 days later. c, Gene-set enrichment analysis reveals the under-representation of cholesterol biosynthesis genes in Raptor-deficient Tregs. The lower part of the plot shows the distribution of the genes in the cholesterol biosynthesis signature gene set (‘Hits’) against the ranked list of genes, and the list on the right shows the top hit genes. d, Real-time PCR quantification of lipogenic gene expression in wild-type and Cd4creRaptorfl/fl freshly isolated Tregs or those stimulated with anti-CD3, anti-CD28 and IL-2 for 8 hours. e, De novo lipogenesis of Tregs from wild-type and Cd4creRaptorfl/fl mice after 24 hours of anti-CD3, anti-CD28 and IL-2 stimulation, with the final 4 hours labeled with 14C-glucose. f, Total cholesterol level was measured using lysates of activated Tregs from wild-type and Cd4creRaptorfl/fl mice. g, In vitro suppression assays mediated by Tregs previously activated by anti-CD3, anti-CD28 and IL-2 for 3 days in the presence of vehicle, simvastatin (2 μM), simvastatin and mevalonate (100 μM) or mevalonate alone. Error bars represent s.d. (n=3). Results represent 2 independent experiments except for c (n=4 from one experiment).