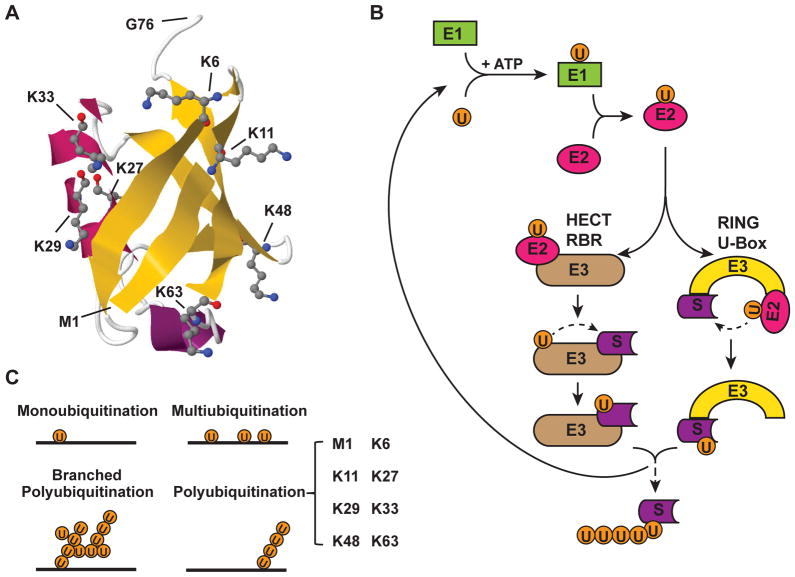
Figure 1. The process of ubiquitination and forms of polyubiquitination.
(A) Structure of a single ubiquitin moiety (Protein Data Bank identifier: 1UBQ) [164] is shown in cartoon style. Secondary structure β-sheet is shown in yellow and α-helix is in magenta. The seven Lys residues and Met1, which can form polyubiquitin linkages with Gly76, are shown in stick-and-ball representation. (B) Ubiquitination is a three-step process which is initiated by activation of ubiquitin. Ubiquitin-activating enzyme (E1) binds to ubiquitin with a high-energy thioester linkage between C-terminus of ubiquitin and active site cysteine residue on the E1, which consumes ATP. Then, the activated ubiquitin moiety is transferred to an ubiquitin-conjugating enzyme (E2) via formation of a second thioester linkage. Finally, with the facilitation of an ubiquitin ligase (E3), ubiquitin is conjugated onto a substrate protein, mostly via an isopeptide bond between the C-terminal Gly76 of the ubiquitin and a internal lysine of the substrate protein. The HECT and RBR family of E3s transfer ubiquitin to substrate via a intermediate step, which involves temporary conjugation of ubiquitin moiety on a internal Cys residue of HECT/RBR E3s. In contrast, the RING and U-box family of E3s modulate direct conjugation of ubiquitin to Lys residue of the substrate from the bound E2. This three-step process can be repeated in multiple cycles, leading to extension of polyubiquitin chain. (C) Depending on the length and number of ubiquitin moiety conjugated on substrate proteins, ubiquitination can be further defined as monoubiquitination (a single ubiquitin moiety conjugated on a single Lys residue of a substrate), multiubiquitination (a single ubiquitin moiety conjugated on multiple Lys residues of a substrate) and polyubiquitination (extended ubiquitin chain forming on a Lys residue of a substrate). Polyubiquitin chain can be linked with homogenous linkages (8 types of linkage as shown in bracket) or with heterogeneous/mixed linkages (also called branched polyubiquitin chain).