Abstract
A widespread downregulated expression of microRNAs (miRNAs) is commonly observed in human cancers. Similarly, deregulated expression of miRNA-processing pathway components, which results in the reduction of global miRNA expression, may also be associated with tumorigenesis. Here, we show that specific ablation of Dicer1 in intestinal epithelial cells accelerates intestinal inflammation-associated tumorigenesis. This effect was apparent only when a single copy of Dicer1 was deleted, but not with complete Dicer1 ablation. DICER expression and subsequent mature miRNA levels were inversely correlated with the number of intact Dicer1 alleles. Because the expression levels of DICER were retained in tumors and its surrounding tissues even after induction of colitis-associated tumors, the effects of Dicer1 deletion were cell-autonomous. Although the expression levels of representative oncogenes and tumor suppressor genes were in most cases inversely correlated with the expression levels of DICER, some genes were not affected by Dicer1 deletion. Thus, deregulating the delicate balance between the expression levels of tumor-promoting and -suppressive genes may be crucial for tumorigenesis in this unique haploinsufficient case.
Introduction
While microRNAs (miRNAs) can function as both tumor suppressors and oncogenes in tumor development [1]–[3], widespread downregulated expression of miRNAs promotes cellular transformation and tumorigenesis and is commonly observed in human cancers [1], [4], [5]. Similar to miRNAs, deregulated expression of miRNA processing pathway components can potentially modify miRNA expression profiles and may be associated with subsequent tumorigenesis and tumor prognosis [6]–[11].
Dicer is a ribonuclease (RNase) III enzyme required for pre-miRNA processing that originates from endogenous single-stranded hairpin- or repeat-associated precursors [12]. A defect in Dicer is one possible mechanism of global downregulated expression of mature miRNAs. Mutations in Dicer were identified in pediatric tumor pleuropulmonary blastoma, Sertoli-Leydig cell tumors, or other tumors [13], [14] and downregulation of DICER is associated with poor prognosis of ovarian cancers and other tumors [11], [15], [16]. These clinical observations, together with experimental results showing the global loss of mature miRNAs induced by Dicer knockdown in vitro and in vivo, demonstrates that Dicer is functionally relevant to oncogenesis [17].
Subsequent studies using mouse models with tissue-specific Dicer1 ablation reported that a single allele of Dicer1 in mice leads to oncogenesis in a lung cancer model caused by a K-ras protooncogene mutation [18], a retinoblastoma model caused by mutations in the tumor suppressor RB gene [19], and a lymphoma model caused by Eµ-myc [20]. In these models, monoallelic, but not complete, loss of Dicer alleles enhanced tumor formation. In addition, although frequent deletion of a Dicer allele has been reported, inactivation of the second wild-type allele has not [18].
Therefore, it has been proposed that Dicer may be unique among classical haploinsufficient tumor suppressor genes since only partial, but not complete, loss promotes tumor development [21]. However, it remains possible that this unique haploinsufficient function of Dicer may be tissue-specific and related to tissue-specific expression of miRNAs and miRNA processing pathway-related genes [20]. It is also unknown whether previous results were dependent on tumorigenesis models caused by enforced mutations in oncogenes or tumor suppressor genes.
To clarify the role of Dicer, we examined mice with intestinal epithelial cell-specific Dicer1 ablation in colitis-associated tumorigenesis. We were particularly interested in the possibility that different effects may result from monoallelic and biallelic ablation of Dicer1.
Results
Intestinal epithelial cell-specific Dicer1-mutant mice
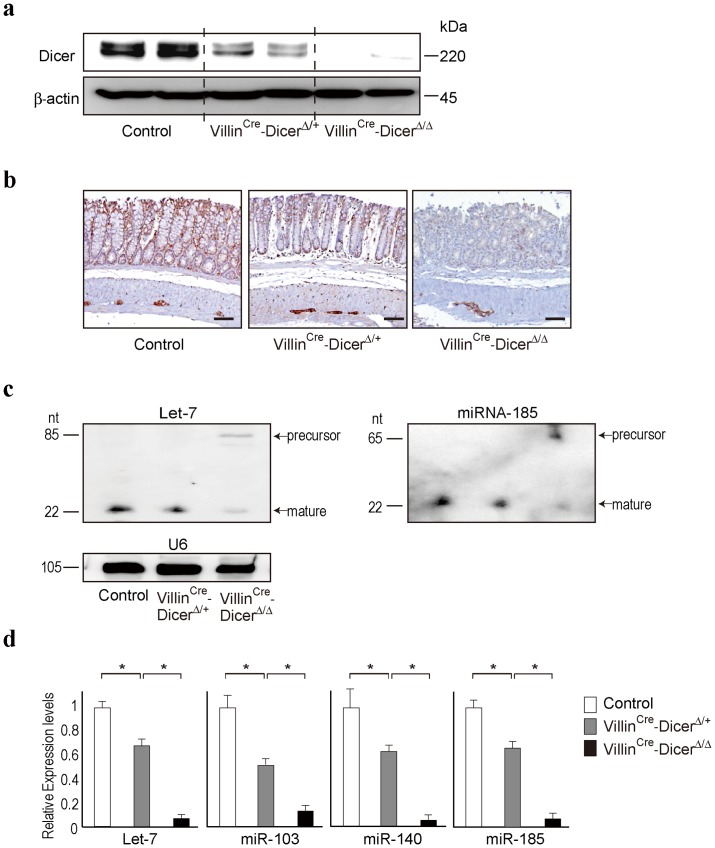
To determine the function of Dicer and its downstream miRNAs in intestinal epithelia, we generated mutant mice lacking Dicer specifically in intestinal epithelial cells (VillinCre-DicerΔ/Δ) by crossing loxP-flanked (Dicerfl/fl) mice with villin promoter driven-Cre (VillinCre)-expressing mice. As comparisons, we used Dicerfl/fl control mice and also included VillinCre-DicerΔ/+ heterozygous mice. While residual DICER protein may be derived cells surrounding the intestinal epithelium that were not completely removed during cell isolations, we confirmed dramatically reduced expression levels of DICER protein in the isolated colonic epithelial cells of VillinCre-DicerΔ/Δ mice (Figure 1a). In contrast, the expression levels of DICER in colonic epithelial cells of VillinCre-DicerΔ/+ heterozygous mice were intermediate to those of Dicerfl/fl control and VillinCre-DicerΔ/Δ mice (Figure 1a). Immunohistochemical analyses of DICER expression using colon tissues of control and Dicer1-mutant mice showed similar DICER expression levels in epithelial cells (Figure 1b). The functional levels of DICER, determined by the abundance of representative mature miRNAs in the colon epithelia of control and mutant mice, appeared to proportionally reflect the deletion of Dicer1 alleles (Figure 1c and d). These results suggest that Dicer was successfully ablated in the intestinal epithelial cells of VillinCre-DicerΔ/Δ homozygous and VillinCre-DicerΔ/+ heterozygous mice, and also suggest that the expression levels of DICER and subsequent function of miRNA maturation were proportionally dependent on the number of mutant alleles.
Figure 1. Intestinal-epithelial-cell-specific Dicer gene ablation.
a, Dicer protein expression levels in the isolated primary colonic epithelial cells of Dicerfl/fl control, VillinCre-DicerΔ/+, and VillinCre-DicerΔ/Δ mice. Representative western blotting images of three independent experiments are shown. b, Representative immunohistochemical images of DICER protein expression (in brown) in the colons of control and mutant mice. The sections are 30 mm proximal to the anal canal. Scale bars = 200 µm. Similar results were obtained from at least three independent mice per group. c, d, The expression levels of the indicated miRNAs in isolated colonic epithelial cells of control and mutant mice determined by Northern blotting (c) and qRT-PCR (d). Representative images of three independent experiments are shown (c). U6 was used as the loading control. The data were obtained from three independent experiments and are shown as means ± s.d. after adjusting the value of control mice as 1. *, p<0.05 (d).
Histological changes of the intestine in intestinal epithelia-specific Dicer1-mutant mice
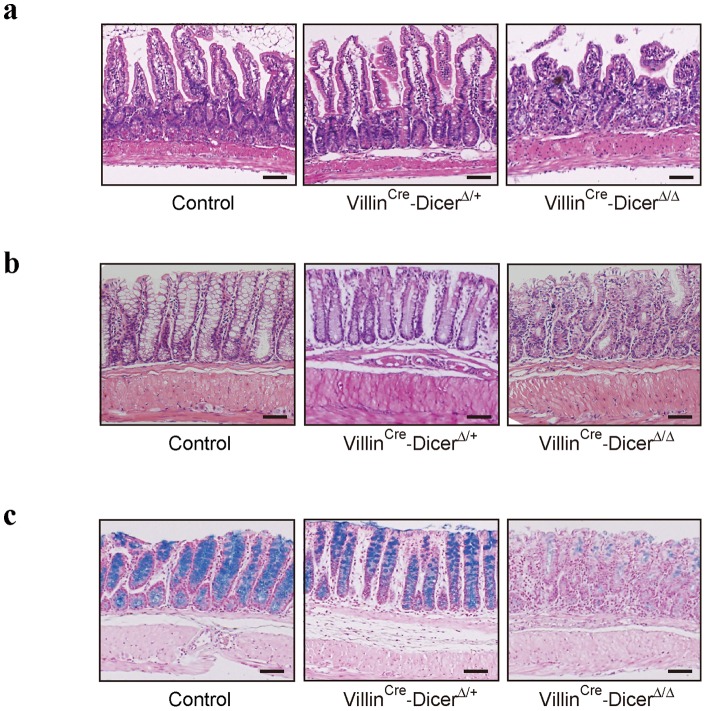
The morphological changes of the intestine in Dicer1-mutant mice were determined using intestinal tissues derived from 12 week old animals. Consistent with a previous report [22], disorganized morphologies of the small intestine lamina propria in Dicer1-mutant mice were clearly observed (Figure 2a). In the large intestine, the Dicer1-mutant mice had disorganized crypt structure (Figure 2b). These deregulated phenomena were most remarkable in VillinCre-DicerΔ/Δ homozygous mice. We also observed increased inflammatory cell infiltration in the lamina propria in Dicer1-mutant mice that was most remarkable in VillinCre-DicerΔ/Δ homozygous mice (Figure 2b and Figure S1). Furthermore, the degree of inflammatory cell infiltration was less in VillinCre-DicerΔ/+ heterozygous mice than in homozygous mice, but more than in Dicerfl/fl control mice (Figure 2b). While Dicer1-mutant mice had fewer goblet cells (Figure 2c), consistent with previous reports [22], [23], VillinCre-DicerΔ/+ heterozygous mice showed an intermediate number of goblet cells in control and homozygous mice (Figure 2c). These results suggest that histological changes in intestinal morphology caused by Dicer mutants are dependent on the expression levels of DICER protein and subsequent expression levels of mature miRNAs.
Figure 2. Phenotypic changes in the intestine of untreated VillinCre-DicerΔ/Δ mice.
a, Hematoxylin and eosin staining of small intestine tissues 40 mm proximal to the ileo-colonic junction. VillinCre-DicerΔ/Δ mice displayes the most deregulated crypt morphologies. Scale bars = 200 μm. Similar results were obtained from at least five mice per group. b, c, Hematoxylin and eosin staining (b) and alcian blue staining (c) of colonic tissues 30 mm proximal to the anal canal. VillinCre-DicerΔ/Δ mice displayes the most disorganized lamina propria structure with increased inflammatory cell infiltration and the fewest goblet cells. Scale bars = 200 μm. Similar results were obtained from at least five mice per group.
Comparable severity of experimental inflammation in the colon of Dicer1-mutant mice
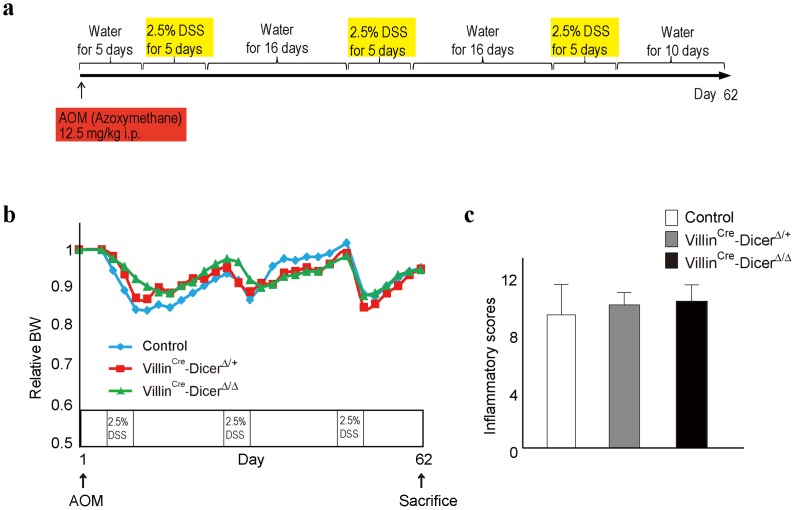
To determine the biological effects of DICER and mature miRNAs in the intestinal epithelial cells on chronic colitis-induced tumorigenesis, we treated eight 12 week-old mice with a single dose of azoxymethane (AOM, 12.5 mg/kg) followed by three cycles of DSS administration in the drinking water (Figure 3a) as a tumor model [24]. AOM is a procarcinogen, which upon metabolic activation causes formation of O6-methyl-guanine [25]. Repeated DSS administration causes chronic inflammation, which greatly enhances the incidence of AOM-induced tumors [24]. During the process of repeated inflammation induction, we examined if the severity of inflammation in the intestine differed among Dicer1-mutant mice. The severity of induced inflammation was determined by the degree of body weight loss, the colon length at the end point, and the histological inflammation scores after one round of DSS treatment, all of which are commonly used to measure the severity of inflammation in this model [26]. Although infiltration of inflammatory cells into the epithelia were observed before induction of experimental inflammation in VillinCre-DicerΔ/Δ homozygous mice, the severity of inflammation after the induction in this model was comparable in control and Dicer1-mutant mice (Figure 3b, c and Figure S2). These results suggested that the inflammation-inducing model used here was severe enough to result in similar inflammation severities in the intestine irrespective of the mutant types.
Figure 3. Comparable severity of inflammation in the colon of VillinCre-DicerΔ/Δ mice.
a, The protocol for experimental inflammation-associated colon tumorigenesis used in this study. b, Body weight changes of the Dicerfl/fl control mice (n = 8), VillinCre-DicerΔ/+ mice (n = 8), or VillinCre-DicerΔ/Δ mice (n = 8) during DSS-induced colon inflammation. Changes in body weight were measured every 2 days. c, Inflammatory scores of mice in each group (n = 8 per group) at day 62 are shown. Data are presented as means ± s.d.
Dicer heterozygous mice were most liable to colitis-associated tumors
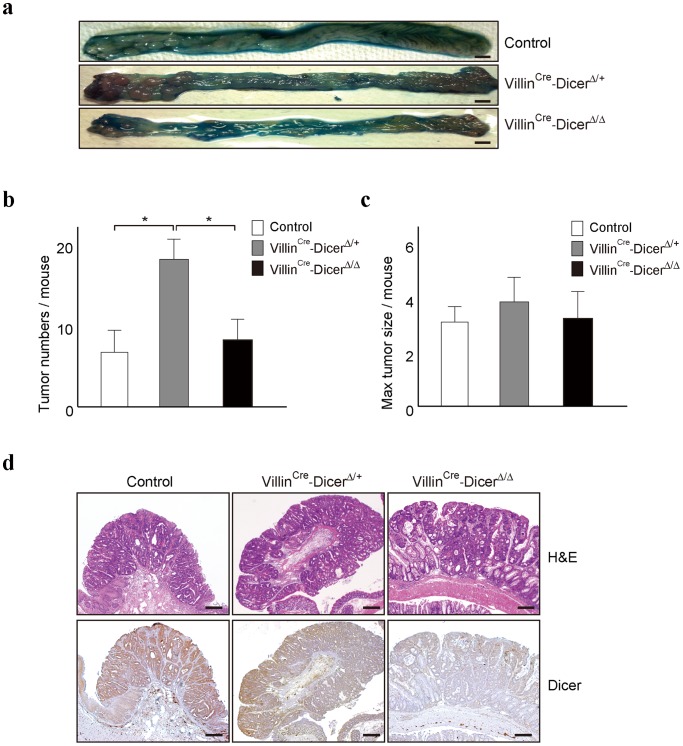
While global down-regulation of miRNAs in many tumors have been reported [1], [4], [27], it was also reported that heterozygosity for the Dicer1 allele, but not complete loss of Dicer1 alleles, enhances tumor development in K-ras-driven lung cancer and RB-driven retinoblastoma models [18], [19]. To determine whether this unique haploinsufficiency concept is applicable to inflammation-associated tumors in the intestine, we compared the number of colon tumors in the AOM-DSS colitis-associated tumor models in three mouse groups (Dicerfl/fl control mice, VillinCre-DicerΔ/+ heterozygous mice, and VillinCre-DicerΔ/Δ homozygous mice). The number of colon tumors at day 62 was significantly greater in VillinCre-DicerΔ/+ heterozygous mice. That is, while seven or eight tumors per mouse in average were observed in Dicerfl/fl control mice or VillinCre-DicerΔ/Δ homozygous mice, more than fifteen tumors per mouse were observed in VillinCre-DicerΔ/+ heterozygous mice (Figure 4a and b). Because inflammation severities were almost similar irrespective of mutant type, the differences in tumor numbers observed here were likely not due to differences in the degree of inflammation. In addition, maximum tumor size was not significantly different (Figure 4c) and the histological tumor appearances were also not clearly different in these three groups (Figure 4d). These results suggest that the unique haploinsufficiency of Dicer is also applicable to inflammation-associated colon tumorigenesis.
Figure 4. Heterozygous mice were more liable to colitis-associated tumors.
a, b, Increased number of tumors in VillinCre-DicerΔ/+ mice. Representative gross colon images are shown in (a). The mean number of tumors per mouse was calculated. Data are presented as means ± s.d. (n = 8 per group) in (b). *, p<0.05. c, The maximum tumor size per mouse was measured. Data are presented as means ± s.d. (n = 8 per group). d, Hematoxylin and eosin staining of colon tumors (upper panels) and immunohistochemical analyses of DICER expression in similar sections of tumors (lower panels) from control and Dicer1-mutant mice after induction of colitis-associated tumors. Scale bars = 200 μm. Representative images are shown from three independent mice per group.
To examine the possibility of non-autonomous effects of DICER on inflammation associated-tumorigenesis and to confirm the expression levels of DICER in the resultant tumors, the expression levels of DICER protein in tumors were determined by immunohistochemistry. The expression levels of DICER protein in tumors were approximately proportional to the number of remaining wild-type Dicer1 alleles and similar to those in the non-tumor colonic epithelial cells (Figure 4d and Figure S3). These results suggest that the effects of DICER expression levels on inflammation-associated tumorigenesis are likely cell-autonomous.
Expression levels of tumor-related genes are associated with DICER expression levels
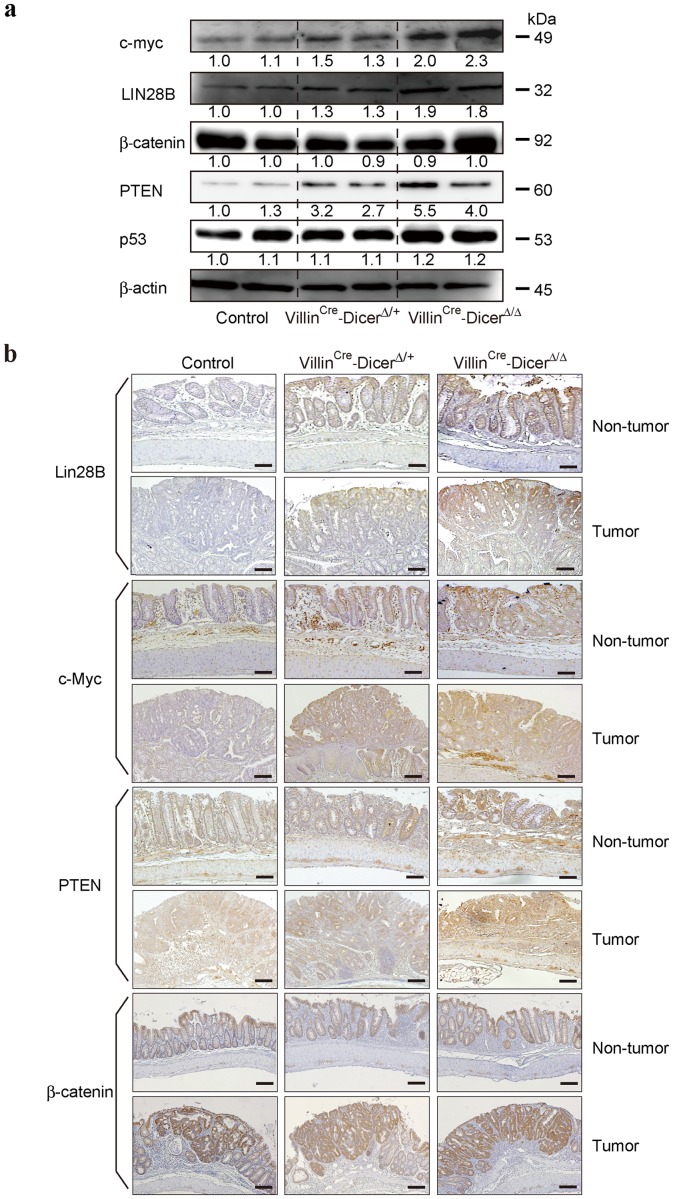
To gain insights into the genes responsible for this Dicer haploinsufficiency in colitis-associated tumorigenesis, we determined the expression levels of representative genes known as oncogenes or tumor suppressors in colon tumorigenesis. Similar to the patterns of DICER protein expression levels in Dicer1 mutants, the expression levels of LIN28B and c-myc, known as oncogenes in colon tumors [28]–[30] regulated by miRNAs [29], [31] were inversely related with the number of wild-type Dicer alleles in both tumors and non-tumor parts of the colonic epithelia (Figure 5a and b and Figure S3). The expression levels of PTEN, a known tumor suppressor gene in the colon, were also dependent on the number of intact Dicer1 alleles in both tumors and non-tumor tissues (Figure 5a and b and Figure S3). However, the expression levels of β-catenin and p53 were not dramatically changed in the Dicer mutants used in this study (Figure 5a and b and Figure S3). These results suggest that the expression levels of most oncogenes and tumor suppressor genes are proportional to DICER expression levels in both tumors and non-tumor tissues, but the magnitude of expression level change is gene-specific.
Figure 5. Expression levels of representative genes in colitis-associated tumors in Dicer1-mutant mice.
a, Western blotting for the indicated genes in isolated colon epithelial cells in bulk after induction of colitis-associated tumors. Representative images of two independent mouse sets are shown. The band intensities were quantitated and adjusted by the expression levels of β-actin. The calculated ratios are indicated below each panel after setting the value of the control mouse as 1.0. b, Immunohistochemical analyses of the indicated protein expression in the tumor and surrounding non-tumor tissue from control and Dicer1-mutant mice after induction of colitis-associated tumors. Scale bars = 200 μm. Representative images are shown. Similar results were obtained from four independent mice per group.
Discussion
In this study, we report the effects of Dicer deletion on intestinal epithelia using intestinal-epithelial-cell-specific Dicer1 ablated mice. In untreated mice, DICER in the intestinal epithelial cells is crucial for the maintenance of mucosal morphology in the small intestine and the differentiation of goblet cells in the large intestine, consistent with a previous report [22], [23]. These observed phenotypes were most severe in homozygous VillinCre-DicerΔ/Δ mice. However, heterozygous VillinCre-DicerΔ/+ mice were most prone to tumors, while Dicer1-null mice developed marginal tumorigenesis, suggesting that the promotion of tumorigenesis is conditional based on the number of wild-type Dicer1 alleles.
Global down-regulation of miRNAs is frequently observed in cancers [1], [4], [5] and deregulation of the miRNA processing pathway is also associated with many types of cancers, which may, in turn, cause aberrations in global miRNA profiles [6]–[11]. MiRNA-processing genes, such as DICER, TRBP and XPO5, are frequently deleted in humans [6]. However, no homozygous deletion of such genes in tumors has been reported and complete loss of Dicer did not preclude tumor formation in in vivo experimental models [21]. Therefore, partial loss of mature miRNAs may be advantageous to tumorigenesis while complete loss may be a disadvantage. From this point of view, partial loss of the expression or functions of miRNA processing pathway molecules, including DICER, may be crucial during tumorigenesis.
The precise mechanisms underlying how monoallelic loss of Dicer leads to greater colitis-associated tumorigenesis are still unknown. Because the severity of inflammation was not significantly different in the model used here, the differences in inflammation severity as a cause of tumorigenesis were not considerable. Cells that have escaped Dicer deletion were the primary source of induced hepatocarcinoma in a liver cancer model [32], which is consistent with less tumorigenesis with a complete loss of Dicer. We hypothesized that Dicer loss may autonomously affect the surrounding cells with residual DICER expression. However, in the colitis-associated tumors examined here, the expression levels of DICER in the tumors as well as in the non-tumor tissues were as expected from the allelic numbers of the disrupted Dicer1. Thus, the effects of Dicer1 disruption were likely cell-autonomous.
The expression levels of representative known tumor suppressors and oncogenes determined in this study were primarily inversely proportional to the DICER expression levels. However, because miRNA regulation of gene expression varies from gene to gene, the impaired delicate balance between oncogenes and tumor suppressive genes may be involved in the unique haploinsufficiency of Dicer in tumorigenesis. While further study is necessary to determine the molecular background of Dicer haploinsufficiency, we consider that the following possibilities should also be elucidated: 1) Genes other than those investigated here are involved in tumorigenesis in this model; 2) DICER may have effects on tumorigenesis through unknown functions other than gene expression regulation through miRNA processing; 3) the accumulated precursors or repetitive RNAs, such as Alu, which could not be processed due to insufficient DICER expression may have effects on the tumorigenesis. These questions should be determined in the future to clarify the unique role of the deregulation of DICER and related deregulation of miRNAs in tumorigenesis.
Untreated Dicer1-mutant mice showed decreased goblet cells and increased inflammatory cell infiltration in the large intestine. DICER and its subsequent miRNA pathway may be related to the differentiation of intestinal epithelial cells, particularly goblet cells [22], [23] and through the deregulation of the epithelial cell differentiation, changes in the microbiome of the gut may lead to infiltration of inflammatory cells into the intestinal mucosa [23]. It cannot be denied that these chronic stress conditions in VillinCre-DicerΔ/Δ mice may also be, to some extent, related to the observed results of Dicer haploinsufficiency in the colitis-associated tumorigenesis.
In conclusion, using intestinal-epithelial-cell-specific Dicer ablated mice, we identified the unique haploinsufficient role of Dicer in colitis-associated tumorigenesis. It is generally accepted that complete loss of tumor suppressor function is necessary for tumor development. However, in this study, monoallelic loss of Dicer in intestinal epithelial cells accelerates tumor formation in colitis-associated tumorigenesis, but complete loss of Dicer did not, which is consistent with previous reports of the role of Dicer in other organs [18]–[20]. These results suggest that complete Dicer inactivation is deleterious for cancer development, while its partial inactivation promotes tumor formation. Although it is clear that the effects of aberrations in the miRNA pathway play a crucial role in tumorigenesis, this unique haploinsufficient role of Dicer needs further analysis to elucidate the intrinsic complex mechanisms and to apply this phenomenon to novel translational applications.
Methods
Mice
Eight-week-old C57BL/6 mice were purchased from CLEA Japan (Tokyo, Japan). VillinCre and Dicerf/f mice were purchased from the Jackson Laboratory (Bar Harbor, ME). Ablation of DICER protein expression was confirmed by antibodies raised against the C-terminal portion of the DICER protein (sc-30226, Santa Cruz Biotechnology, Santa Cruz, CA). All experimental protocols were approved by the Ethics Committee for Animal Experimentation at the Graduate School of Medicine, the University of Tokyo, Japan and conducted in accordance with the Guidelines for the Care and Use of Laboratory Animals of the Department of Medicine, the University of Tokyo.
Inflammation associated-colon tumor model
Mice (8–12 weeks old) were injected intraperitoneally with 12.5 mg/kg azoxymethane (AOM; Wako, Osaka, Japan). After 5 days, 2.5% dextran sulfate sodium (DSS; ICN Biomedicals, Irvine, CA) was mixed in the drinking water for 5 days, followed by 16 days of regular water. This cycle was repeated twice and mice were sacrificed 10 days after the last cycle. Induced colon tumors were counted and sized. Body weight was checked every 2 days. Histological inflammatory scoring of the colon tissues was performed in a blinded manner as reported previously [26]. The macroscopic images were taken after Indigo carmine staining.
Isolation of primary intestinal epithelial cells
Intestinal epithelial cells were isolated using a slightly modified rapid low-temperature method [33]. Briefly, the entire colon was removed and washed with ice-cold PBS. The intestine was divided into 2–3 mm-long fragments and transferred into chelating buffer (27 mM trisodium citrate, 5 mM Na2PO4, 96 mM NaCl, 8 mM KH2PO4, 1.5 mM KCl, 0.5 mM DTT, 55 mM D-sorbitol, 44 mM sucrose, 6 mM EDTA, 5 mM EGTA, pH 7.3) for 45 min at 4°C. Intestinal epithelial cells were dissociated by repeated vigorous shaking. Tissue debris was removed using a 100 µm cell-strainer and cells were collected by centrifugation at 150×g for 10 min at 4°C. The viability of cells was confirmed by trypan blue staining and cells processed for protein extraction.
Quantitative RT-PCR and Northern blotting for miRNAs
Quantitative RT-PCR analysis of miRNA expression was performed as described previously [34]. Northern blotting analysis of miRNAs was performed as described previously [35]. Briefly, total RNA was extracted using TRIzol Reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. Five μg of small RNA were resolved in denaturing 15% polyacrylamide gels containing 7 M urea in 1× TBE and transferred to Hybond N+ membrane (GE Healthcare, Waukesha, WI) in 0.25× TBE. Membranes were UV-crosslinked and prehybridized in hybridization buffer. Hybridization was performed overnight at 42°C in ULTRAhyb-Oligo Buffer (Ambion, Austin, TX) containing a biotinylated probe specific for let-7b and miR-185, which had previously been heated to 95°C for 2 min. Membranes were washed at 42°C in 2× SSC containing 0.1% SDS, and the bound probe was visualized using a BrightStar BioDetect Kit (Ambion). Blots were stripped by boiling in a solution containing 0.1% SDS, 5 mM EDTA for 10 min prior to rehybridization with a U6 probe with the sequence 5′-CACGAATTTGCGTGTCATCCTT′-3.
Western blotting
Western blotting was performed as described previously [36]. The following antibodies were used: anti-DICER (SAB4200087) and anti-β-actin (A5316) were purchased from Sigma-Aldrich (St. Louis, MO). Anti-c-Myc (sc-40) and anti-DICER (sc-30226) were purchased from Santa Cruz Biotechnology. Anti-LIN28B (5422), anti-PTEN (9559), and anti-β-catenin (9582) were purchased from Cell Signaling Technology (Danvers, MA). Anti-p53 (OP03) was purchased from Calbiochem (San Diego, CA).
Immunohistochemistry
Immunohistochemistry was performed as described previously [36]. The following antibodies were used: anti-LIN28B (16178-1-A, Proteintech Group, Chicago, IL), anti-DICER, anti-c-myc, anti-PTEN, and anti-β-catenin described above.
Statistical analysis
Significant differences between groups were determined using Student's t-test when variances were equal. When variances were unequal, Welch's t-test was used. P-values <0.05 were considered to indicate statistical significance.
Supporting Information
Inflammatory scores in the colon of untreated Dicer1-mutant mice. Inflammatory scores in the colon of untreated mice from each group (n = 4 per group). Data are presented as means ± s.d. *, p<0.05.
(PDF)
Severity of inflammation after induction of colitis in Dicer1 -mutant mice. a, Representative colonic tissue images in hematoxylin and eosin staining at day 12. Control and Dicer1-mutant mice showed similar inflammation severity. The sections are 30 mm proximal to the anal canal. Scale bars = 200 μm. Similar results were obtained from five independent mice per group. b, The length of the colon in mice at day 62 is shown. Data are presented as means ± s.d. (n = 8 per group).
(PDF)
Immunohistochemical analyses of protein expression in control and Dicer1 -mutant mice after induction of colitis-associated tumors. Immunohistochemical analyses of the indicated protein expression in tumors and surrounding non-tumor tissue from control and Dicer1-mutant mice after induction of colitis-associated tumors. Scale bars = 200 μm. Dashed lines indicate the border between the tumor and its surrounding tissues. Representative images are shown. Similar results were obtained from four independent mice per group.
(PDF)
Funding Statement
This work was supported by Grants-in-Aid from the Ministry of Education, Culture, Sports, Science and Technology, Japan (#22390058 and #2529307 to M. Otsuka, and #24390183 to K. Koike), by Health Sciences Research Grants of The Ministry of Health, Labour and Welfare of Japan (to K. Koike), grants from the Japan Foundation for Applied Enzymology, the Sagawa Foundation for Promotion of Cancer Research, the Cell Science Research Foundation, and the Japanese Society of Gastroenterology (to M. Otsuka.). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Calin GA, Croce CM (2006) MicroRNA signatures in human cancers. Nat Rev Cancer 6: 857–866. [DOI] [PubMed] [Google Scholar]
- 2. Zhang L, Huang J, Yang N, Greshock J, Megraw MS, et al. (2006) microRNAs exhibit high frequency genomic alterations in human cancer. Proc Natl Acad Sci U S A 103: 9136–9141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Croce CM, Calin GA (2005) miRNAs, cancer, and stem cell division. Cell 122: 6–7. [DOI] [PubMed] [Google Scholar]
- 4.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, et al.. (2005) MicroRNA expression profiles classify human cancers. Nature. England. 834–838. [DOI] [PubMed]
- 5. Gaur A, Jewell DA, Liang Y, Ridzon D, Moore JH, et al. (2007) Characterization of microRNA expression levels and their biological correlates in human cancer cell lines. Cancer Res 67: 2456–2468. [DOI] [PubMed] [Google Scholar]
- 6. Bahubeshi A, Tischkowitz M, Foulkes WD (2011) miRNA processing and human cancer: DICER1 cuts the mustard. Sci Transl Med 3: 111ps146. [DOI] [PubMed] [Google Scholar]
- 7. Heravi-Moussavi A, Anglesio MS, Cheng SW, Senz J, Yang W, et al. (2012) Recurrent somatic DICER1 mutations in nonepithelial ovarian cancers. N Engl J Med 366: 234–242. [DOI] [PubMed] [Google Scholar]
- 8. Martello G, Rosato A, Ferrari F, Manfrin A, Cordenonsi M, et al. (2010) A MicroRNA targeting dicer for metastasis control. Cell 141: 1195–1207. [DOI] [PubMed] [Google Scholar]
- 9.Melo SA, Ropero S, Moutinho C, Aaltonen LA, Yamamoto H, et al.. (2009) A TARBP2 mutation in human cancer impairs microRNA processing and DICER1 function. Nat Genet. United States. 365–370. [DOI] [PMC free article] [PubMed] [Retracted]
- 10. Melo SA, Moutinho C, Ropero S, Calin GA, Rossi S, et al. (2010) A genetic defect in exportin-5 traps precursor microRNAs in the nucleus of cancer cells. Cancer Cell 18: 303–315. [DOI] [PubMed] [Google Scholar]
- 11. Merritt WM, Lin YG, Han LY, Kamat AA, Spannuth WA, et al. (2008) Dicer, Drosha, and outcomes in patients with ovarian cancer. N Engl J Med 359: 2641–2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bartel DP (2009) MicroRNAs: target recognition and regulatory functions. Cell 136: 215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hill DA, Ivanovich J, Priest JR, Gurnett CA, Dehner LP, et al. (2009) DICER1 mutations in familial pleuropulmonary blastoma. Science 325: 965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rio Frio T, Bahubeshi A, Kanellopoulou C, Hamel N, Niedziela M, et al. (2011) DICER1 mutations in familial multinodular goiter with and without ovarian Sertoli-Leydig cell tumors. JAMA 305: 68–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kitagawa N, Ojima H, Shirakihara T, Shimizu H, Kokubu A, et al. (2013) Downregulation of the microRNA biogenesis components and its association with poor prognosis in hepatocellular carcinoma. Cancer Sci 104: 543–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhu DX, Fan L, Lu RN, Fang C, Shen WY, et al. (2012) Downregulated Dicer expression predicts poor prognosis in chronic lymphocytic leukemia. Cancer Sci 103: 875–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T (2007) Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet 39: 673–677. [DOI] [PubMed] [Google Scholar]
- 18. Kumar MS, Pester RE, Chen CY, Lane K, Chin C, et al. (2009) Dicer1 functions as a haploinsufficient tumor suppressor. Genes Dev 23: 2700–2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lambertz I, Nittner D, Mestdagh P, Denecker G, Vandesompele J, et al. (2010) Monoallelic but not biallelic loss of Dicer1 promotes tumorigenesis in vivo. Cell Death Differ 17: 633–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Arrate MP, Vincent T, Odvody J, Kar R, Jones SN, et al. (2010) MicroRNA biogenesis is required for Myc-induced B-cell lymphoma development and survival. Cancer Res 70: 6083–6092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Davalos V, Esteller M (2012) Rolling the dice to discover the role of DICER in tumorigenesis. Cancer Cell 21: 717–719. [DOI] [PubMed] [Google Scholar]
- 22.McKenna LB, Schug J, Vourekas A, McKenna JB, Bramswig NC, et al.. (2010) MicroRNAs control intestinal epithelial differentiation, architecture, and barrier function. Gastroenterology 139: 1654–1664, 1664.e1651. [DOI] [PMC free article] [PubMed]
- 23. Biton M, Levin A, Slyper M, Alkalay I, Horwitz E, et al. (2011) Epithelial microRNAs regulate gut mucosal immunity via epithelium-T cell crosstalk. Nat Immunol 12: 239–246. [DOI] [PubMed] [Google Scholar]
- 24. Okayasu I, Ohkusa T, Kajiura K, Kanno J, Sakamoto S (1996) Promotion of colorectal neoplasia in experimental murine ulcerative colitis. Gut 39: 87–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pegg AE (1984) Methylation of the O6 position of guanine in DNA is the most likely initiating event in carcinogenesis by methylating agents. Cancer Invest 2: 223–231. [DOI] [PubMed] [Google Scholar]
- 26.Otsuka M, Kang YJ, Ren J, Jiang H, Wang Y, et al.. (2010) Distinct effects of p38alpha deletion in myeloid lineage and gut epithelia in mouse models of inflammatory bowel disease. Gastroenterology 138: 1255–1265, 1265.e1251–1259. [DOI] [PMC free article] [PubMed]
- 27. Ventura A, Jacks T (2009) MicroRNAs and cancer: short RNAs go a long way. Cell 136: 586–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. King CE, Cuatrecasas M, Castells A, Sepulveda AR, Lee JS, et al. (2011) LIN28B promotes colon cancer progression and metastasis. Cancer Res 71: 4260–4268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. King CE, Wang L, Winograd R, Madison BB, Mongroo PS, et al. (2011) LIN28B fosters colon cancer migration, invasion and transformation through let-7-dependent and -independent mechanisms. Oncogene 30: 4185–4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Network CGA (2012) Comprehensive molecular characterization of human colon and rectal cancer. Nature 487: 330–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Melton C, Judson RL, Blelloch R (2010) Opposing microRNA families regulate self-renewal in mouse embryonic stem cells. Nature 463: 621–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sekine S, Ogawa R, Ito R, Hiraoka N, McManus MT, et al.. (2009) Disruption of Dicer1 induces dysregulated fetal gene expression and promotes hepatocarcinogenesis. Gastroenterology. United States. 2304–2315 e2301–2304. [DOI] [PMC free article] [PubMed]
- 33. Flint N, Cove FL, Evans GS (1991) A low-temperature method for the isolation of small-intestinal epithelium along the crypt-villus axis. Biochem J 280 ( Pt 2): 331–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yoshikawa T, Takata A, Otsuka M, Kishikawa T, Kojima K, et al.. (2012) Silencing of microRNA-122 enhances interferon-α signaling in the liver through regulating SOCS3 promoter methylation. Sci Rep 2. [DOI] [PMC free article] [PubMed]
- 35. Takata A, Otsuka M, Yoshikawa T, Kishikawa T, Hikiba Y, et al. (2013) MicroRNA-140 acts as a liver tumor suppressor by controlling NF-κB activity by directly targeting DNA methyltransferase 1 (Dnmt1) expression. Hepatology 57: 162–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kojima K, Takata A, Vadnais C, Otsuka M, Yoshikawa T, et al. (2011) MicroRNA122 is a key regulator of α-fetoprotein expression and influences the aggressiveness of hepatocellular carcinoma. Nat Commun 2: 338. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Inflammatory scores in the colon of untreated Dicer1-mutant mice. Inflammatory scores in the colon of untreated mice from each group (n = 4 per group). Data are presented as means ± s.d. *, p<0.05.
(PDF)
Severity of inflammation after induction of colitis in Dicer1 -mutant mice. a, Representative colonic tissue images in hematoxylin and eosin staining at day 12. Control and Dicer1-mutant mice showed similar inflammation severity. The sections are 30 mm proximal to the anal canal. Scale bars = 200 μm. Similar results were obtained from five independent mice per group. b, The length of the colon in mice at day 62 is shown. Data are presented as means ± s.d. (n = 8 per group).
(PDF)
Immunohistochemical analyses of protein expression in control and Dicer1 -mutant mice after induction of colitis-associated tumors. Immunohistochemical analyses of the indicated protein expression in tumors and surrounding non-tumor tissue from control and Dicer1-mutant mice after induction of colitis-associated tumors. Scale bars = 200 μm. Dashed lines indicate the border between the tumor and its surrounding tissues. Representative images are shown. Similar results were obtained from four independent mice per group.
(PDF)