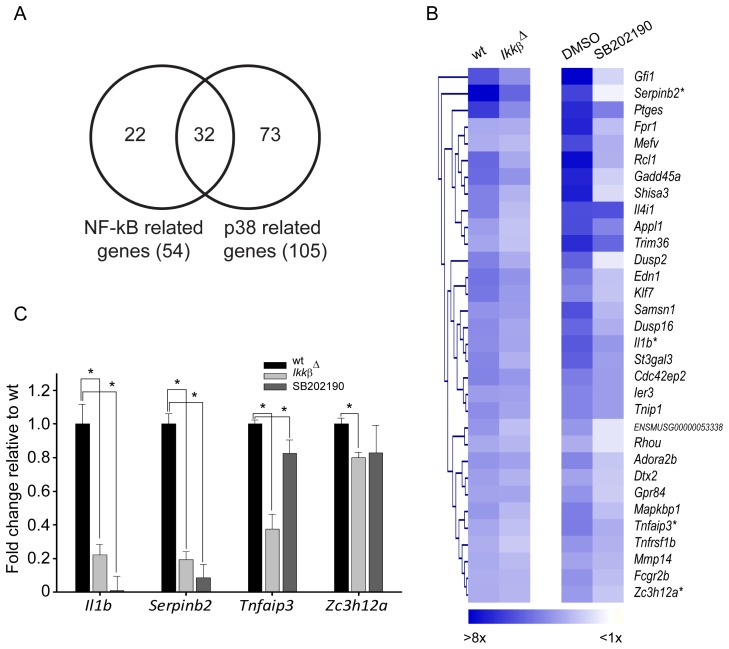
Figure 2. Identification of LPS-induced genes that were regulated by both NF-κB and p38.
(A) Venn diagram of NF-κB and p38-dependent genes. NF-κB-related genes were identified from genes that were down-regulated in Ikkβ Δ BMDMs as compared with wt BMDMs after LPS treatment, and p38-related genes were selected by comparing SB202190- (p38-inhibitor) with dimethyl sulfoxide (DMSO)-treated BMDMs after LPS treatment. Thirty-two LPS-induced genes were regulated by both NF-κB and p38-downstream transcription factors. (B) Hierarchical clustering of average fold change for the NF-κB and p38-dependent genes. Each column represents the average fold change 4 h after LPS treatment compared to 0 h. *: genes chosen for PCR validation. (C) Relative fold changes of Il1b, Serpinb2, Tnfaip3, and Zc3h12a mRNA from BMDMs from wt and Ikkβ Δ cells stimulated with LPS (100 ng/mL) for 4 h in the presence or absence of SB202190 (10 µM) were measured by quantitative RT-PCR. The fold change of 4 h vs. 0 h was normalized to wt BMDMs. The internal control was Cyclophilin A mRNA (Cypa). Data represent the mean ± SD for at least two independent experiments. *, P<0.05.