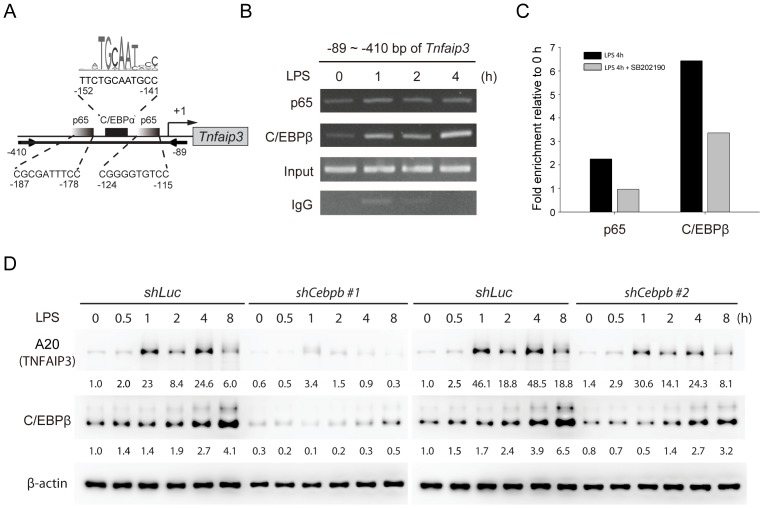
Figure 4. NF-κB and C/EBPβ are required for induction of Tnfaip3 in LPS-activated macrophages.
(A) Schematic map of predicted NF-κB p65 and C/EBPβ DNA binding sites in the promoter of Tnfaip3. oPOSSUM (http://www.cisreg.ca/oPOSSUM/) was used as the prediction tool. The JASPAR CORE vertebrate database was selected for transcription factor binding site matrices. Two arrows depict the PCR primers. (B) p65 and C/EBPβ bind to the promoter of Tnfaip3 after LPS stimulation. RAW264.7 cells were treated with LPS (100 ng/ml) for the indicated times. Chromatin was immunoprecipitated with anti-p65 and anti-C/EBPβ antibodies. Rabbit IgG was a negative control. Precipitated DNA or 1% of the chromatin input was amplified with primers for the Tnfaip3 promoter (−89 ∼ −410). The PCR products were loaded and separated on a 2% agarose gel. One of two independent experiments is shown. (C) LPS-induced association of p65 and C/EBPβ with Tnfaip3 was reduced in the presence of p38 inhibition. Chromatin isolated from RAW264.7 cells treated with LPS (100 ng/ml) for 4 h in the absence or presence of SB202190 (10 µM) were subjected to ChIP assay as described above. The relative quantity of promoter enriched by ChIP was quantified by real-time PCR and expressed as the fold enrichment of untreated control samples after normalization to rabbit IgG. Data represent the means of two independent experiments. (D) LPS-induced expression of A20 (TNFAIP3) was decreased in C/EBPβ-depleted RAW264.7 cells. Cells were infected with lentiviruses encoding shRNA against luciferase (shLuc) or C/EBPβ (shCebpb), and treated with LPS (100 ng/ml) for the indicated times. Cell lysates were collected and analyzed by immunoblotting using the indicated antibodies.