Figure 1.
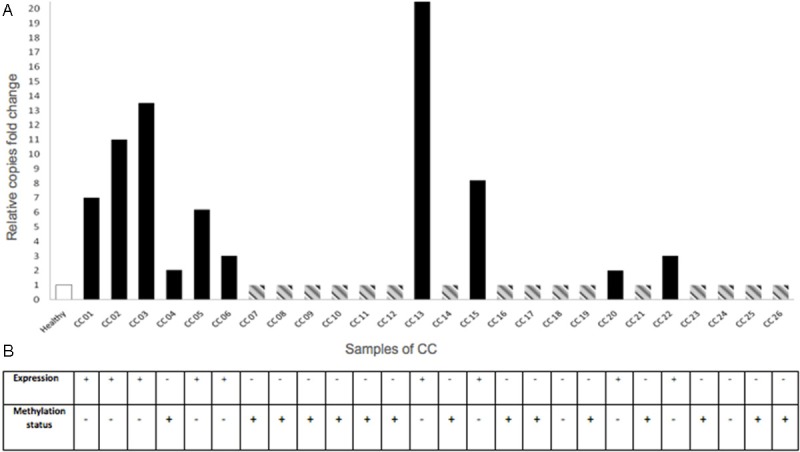
Molecular events for CRBP1 gene in cervical epithelium samples. A: In order to know the gain of copy number of the CRBP1 gene, DNA of healthy cervix and CC samples, were subjected to real time PCR with specific Taqman probes. White bar (healthy cervix samples) represents the mean of the normal cervices (n = 26) without extra copies of CRBP1 gene. Black bars show CC samples with gain of copy number (2-20X); while gray dotted line bars are showing CC samples that do not change in the copies number. Values above the cut-off line (as 1), being assigned as increased gene copy number compared with normal cervical epithelium. CRBP1 Hs01437985_cn probe, and Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) Hs00894322_cn probe were used as reference; the relative genomic copy number was calculated using the comparative Ct methods [26]. In X-axis represents cervical samples, Y-axis relative copies fold change of CRBP1 gene. B: CRBP1 expression was observed as positive immunostaining result on tissue microarray as mentioned in Methods section The DNAs used for gain of copy number (panel A) were also used for the methylation assay. Methylation result represents the methylation of the CRBP1 promoter. In this case, each healthy or CC sample, correspond to each column for CRBP1 expression and methylation status. Interestingly, in most of the cases, there was an association between the lack of expression of the CRBP1 gene and its methylation status.
