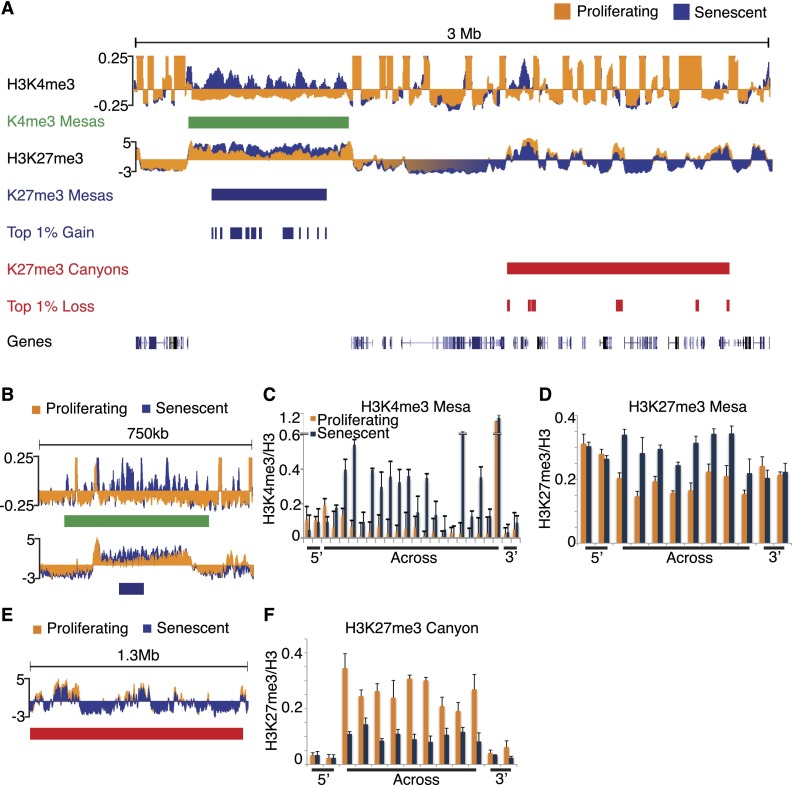
Figure 1.
Large-scale chromatin changes occur in the senescence genome for both histone modifications. (A) Sample tracks of H3K4me3 and H3K27me3 ChIP-seq data over a 3-Mb region of chromosome 1 (Chr1: 38,016,703–41,116,920) show major regions of gains and losses for both histone modifications. Proliferating tracks are shown in orange, and senescence tracks are shown in blue. K4me3 mesas are shown in green blocks, the top 1% gain regions and K27me3 mesas are shown in blue blocks, and the top 1% loss regions and K27me3 canyons are shown in red blocks. (B) Example of overlapped K4me3 and K27me3 mesas over a 750-kb region of chromosome 7 (Chr7: 35,060,526–35,814,473). Proliferating tracks are shown in orange, and senescence tracks are shown in blue. (C,D) ChIP-qPCR validation of the K4me3 (C) and K27me3 (D) mesas shown in B. Proliferating data are shown in orange, and senescence data are shown in blue. ChIP-qPCR primers are tiled across the entire region of the mesa and include 3′ and 5′ flanking primers. ChIP-qPCR data are shown as ratios of modification to total histone H3. ChIP-qPCR data are the average of three biological replicates, and error bars represent standard deviation from the mean. (E) Example of K27me3 canyon over a 1.3-Mb region of chromosome 6 (Chr6: 36,239,850–37,570,928). Proliferating tracks are shown in orange, and senescence tracks are shown in blue. (F) ChIP-qPCR validation of the K27me3 canyon shown in E. Proliferating data are shown in orange, and senescence data are shown in blue. ChIP-qPCR primers are tiled across the entire region of the canyon and include 3′ and 5′ flanking primers. ChIP-qPCR data are shown as ratios of modification to total histone H3. ChIP-qPCR data are the average of three biological replicates, and error bars represent the standard deviation from the mean.