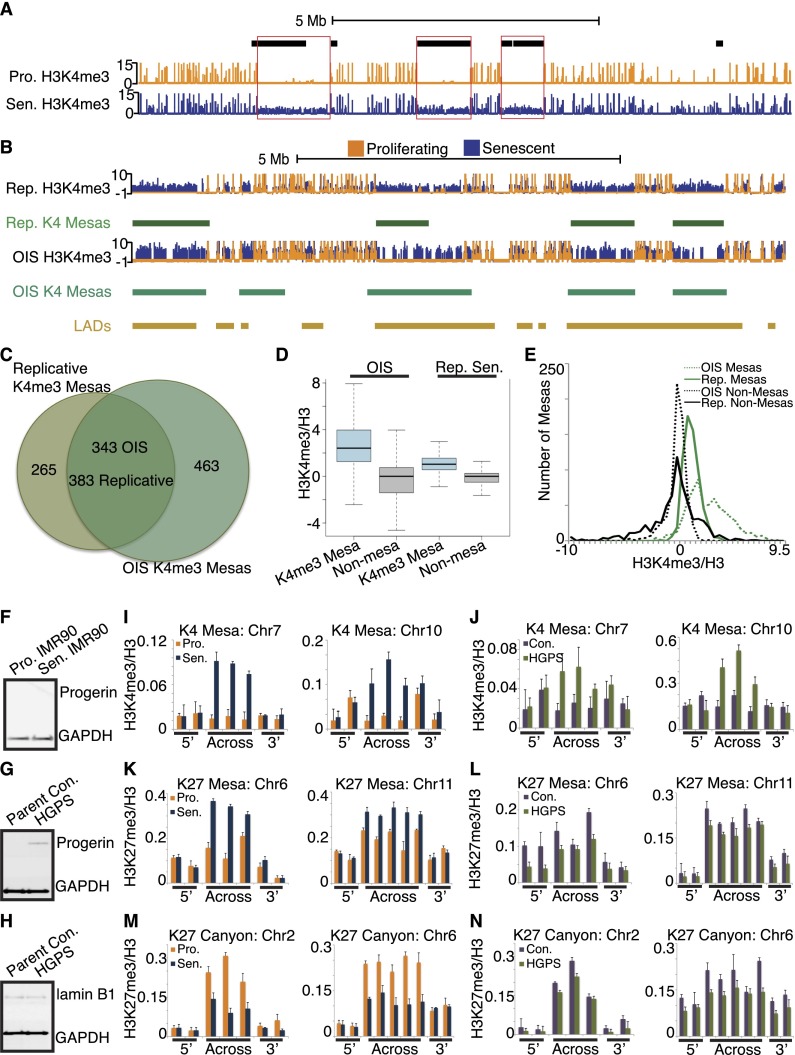
Figure 3.
H3K4me3 mesas overlap hypomethylated regions in cancer; mesas form in an OIS model and HGPS. (A) Example track view of a cluster of H3K4me3 mesas on chromosome 1 (Chr1: 30,500,000–40,000,000) shows a strong degree of overlap with cancer hypomethylated regions (black blocks). Proliferating tracks are shown in orange, and senescence tracks are shown in blue. (B) Example track view of the cluster of H3K4me3 mesas between replicative senescence and OIS shows a strong degree of overlap between the mesas of the two different senescence models. Proliferating tracks are shown in orange, and senescence tracks are shown in blue. Replicative senescence mesas are shown in green blocks, OIS mesas are shown in teal blocks, and LADs are shown in gold blocks. (C) Venn diagram representing the degree of overlap between the OIS and replicative senescence mesas shows a high degree of overlap between the two sets. Note that the OIS model has almost 200 more mesas than the replicative senescence. (D) Box plot analysis of the K4me3 mesas in replicative senescence and OIS (blue boxes) compared with nonmesa control regions (gray) shows an even higher enrichment for H3K4me3 gain in OIS mesas than replicative senescence mesas. (E) Histogram analysis of the OIS and replicative senescence mesas shows a bimodal pattern for the OIS mesas (green dotted line) compared with the single mode of replicative senescence mesas (green solid line). This suggests that a subset of OIS mesas is particularly enriched for H3K4me3, even more than the replicative senescence mesas. Control nonmesa regions are shown in black solid (replicative senescence) and dotted (OIS) lines. (F,G) Western blot analysis of progerin expression in proliferating and senescent IMR90s (F) and parental control and HGPS cell strains (G) shows progerin expression only in HGPS cells, not in IMR90 or parental control cells. (H) Western blot analysis of lamin B1 expression in parental control and HGPS cell strains shows similar levels of lamin B1 in both cell populations at the time of experimentation, underscoring the proliferating state of the HGPS cells at this point. (I,J) ChIP-qPCR evidence for K4me3 mesa formation in HGPS. Replicative senescence qPCR mesa validation shown in I (proliferating shown in orange and senescence shown in blue). The same K4me3 mesa analysis shown in J (parent control shown in purple and HGPS shown in green) indicates that K4me3 mesas may be a shared feature of HGPS. ChIP-qPCR primers are tiled across the region of the mesas and include 3′ and 5′ flanking primers. ChIP-qPCR data are shown as ratios of modification to total histone H3. ChIP-qPCR data are the average of three biological replicates, and error bars represent the standard deviation from the mean. (K,L) ChIP-qPCR test for K27me3 mesa formation in HGPS suggests that these features may not be forming in proliferating HGPS. Replicative senescence qPCR mesa validation shown in K (proliferating shown in orange and senescence shown in blue). The same K4me3 mesa analysis shown in L (parent control shown in purple and HGPS shown in green) indicates that K27me3 mesas may not be a shared feature of HGPS. Note that the K27me3 signal in HGPS is generally lower than the parental control, even at flanking regions. ChIP-qPCR primers are tiled across the region of the mesas and include 3′ and 5′ flanking primers. ChIP-qPCR data are shown as ratios of modification to total histone H3. ChIP-qPCR data are the average of three biological replicates, and error bars represent the standard deviation from the mean. (M,N) ChIP-qPCR test for K27me3 canyon formation in HGPS suggests that these features may not be forming in proliferating HGPS. Replicative senescence qPCR canyon validation shown in M (proliferating shown in orange and senescence shown in blue). The same K4me3 canyon analysis shown in N (parent control shown in purple and HGPS shown in green) indicates that K27me3 canyons are maybe not forming in still-proliferating HGPS cells. Note that the K27me3 signal in HGPS is generally lower than the parental control, even at flanking regions. ChIP-qPCR primers are tiled across the region of the canyons and include 3′ and 5′ flanking primers. ChIP-qPCR data are shown as ratios of modification to total histone H3. ChIP-qPCR data are the average of three biological replicates, and error bars represent the standard deviation from the mean.