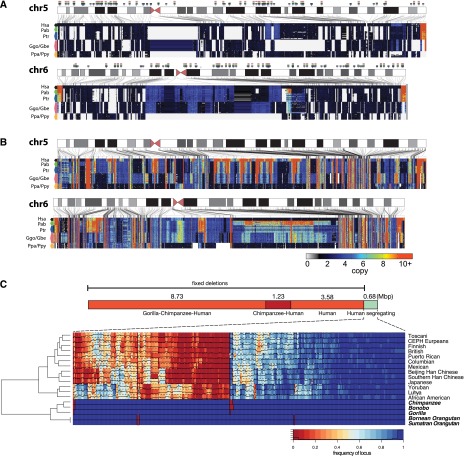
Figure 1.
Duplication and deletion landscape. (A) Ideograms of human autosomes 5 and 6 overlaid with copy number heat maps of the deletion landscape of great apes across seven species and 11 distinct populations. Each row represents one of 97 individuals sorted by species; each column shows the estimated copy number in each of these individuals for deleted loci in 500-bp unmasked windows. Arrows above the chromosome ideogram indicate deletions identified along the lineages leading to the human species, the African great ape, chimpanzee–human, and human lineages, respectively. (B) Ideograms of human autosomes 5 and 6 overlaid with copy number heat maps of the duplication landscape of great apes. (C) Breakdown of the number of base pairs lost along the lineage leading to humans identified by screening sequence absent from the human reference genome yet present in the orangutan, gorilla, or chimpanzee reference genomes against the 97 great apes sequenced in this study. A total of 13.54 Mb has been lost in these lineages since the divergence of African great apes and orangutans. We find that an additional 680 kb (316 loci) of sequence absent from the human reference genome (4.8% of the total) is fixed in all nonhuman great apes and segregating in humans. For these loci a hierarchically clustered heat map is shown. Colors indicate the frequency of sequences absent in the human reference genome assessed in 624 diverse individuals from 13 different populations sequenced to low coverage by the 1000 Genomes Project and found to be segregating with ≥5% frequency in at least one population. The hierarchical clustering recapitulates all the relationships between the individual human populations and the different great ape species assessed in this study. We identify 53.8 kb of sequence segregating exclusively in African populations compared to only 1.4 kb of sequence segregating specifically in Europeans.