Figure 3.
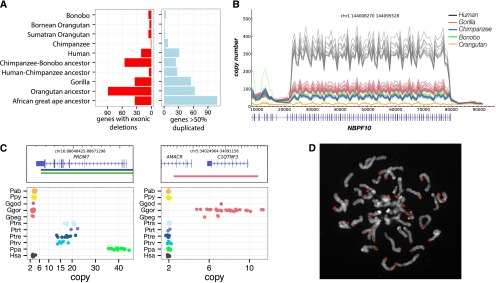
Genic variation. (A) Summary of the number of genes with exonic deletions and genes duplicated in each of the lineages assessed in this study. We identify 340 genes lost throughout the great ape lineage. While orangutans show the highest number of gene-exon-loss events (89), strikingly, the second highest number of gene-exon-loss events was in the chimpanzee–bonobo ancestral lineage, where 55 were lost. (B) A line plot of the copy number over the DUF1220 domain of NBPF10. This domain has expanded specifically in the African great ape lineage with human exhibiting ∼300 copies compared to 50–100 in chimpanzee, bonobo, and gorilla. (C) Lineage-specific great ape duplication events encompassing genes. Gene models are drawn with the duplication breakpoints shown below colored by lineage and dot-plots of the copy number in all individuals assessed in this study. PRDM7 is a close paralog of PRDM9, the binding of which associates with recombination hotspots in humans. We find that PRDM7 is specifically duplicated in the Pan genus and highly stratified; Nigerian–Cameroon chimpanzees have 10–15 copies, while Eastern and Central chimpanzees have 16–20. Bonobos exhibit 30–40 copies of the gene. FISH assays demonstrate the extra copies to be the result of subtelomeric duplicative transposition (D). AMACR and C1QTNF3 are also specifically duplicated in gorillas. Mutations in AMACR have been shown to result in adult-onset neurological disorders (Ferdinandusse et al. 2000), and C1QTNF plays a key role in skeletal development, inducing increased growth of murine mesenchymal cells with overexpression (Maeda et al. 2001).
