Figure 4.
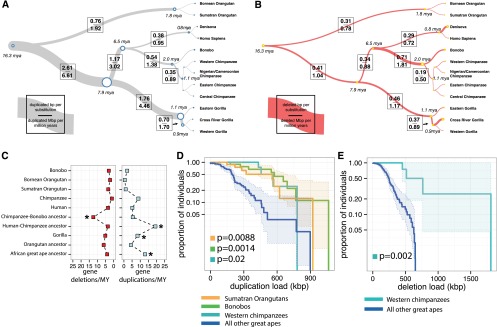
CNV rates and polymorphism. Rates of duplication (A) and deletion accumulation (B) as a function of the number of substitutions along each branch of the great ape phylogeny. Tree branch lengths are scaled proportionally to the number of substitutions, while tree widths are scaled proportionally to the number of duplicated base pairs per substituted base pair. Duplicated base pairs were added ∼2.6-fold the rate of substitution along the African great ape ancestral branch, which rapidly declined in the chimpanzee–human ancestral lineage and more slowly in the gorilla lineage. In contrast, the rate of deletion in the great ape lineage is fairly consistent along all branches (mean of 0.32 deleted base pairs per substitution) with the exception of the chimpanzee–bonobo ancestral lineage, where an approximate twofold increase in the rate of deletion is observed (0.71 deleted base pairs per substitution). (C) The rate of genic deletion events and gene duplication events per million years plotted for each of the lineages assessed in this study. The rate of gene deletion events is significantly higher in the chimpanzee–bonobo ancestor (P = 5.262 × 10−9). An acceleration in the number of gene duplications is observed in the African great ape ancestor, the human–chimpanzee ancestor, and the ancestral gorilla lineage (P = 1.663 × 10−20). (D) A survival curve of the total load of segregating duplications >30 kb in Western chimpanzees, Sumatran orangutans, and bonobos compared to all other great apes shows that these three populations harbor an increased total number of duplicated base pairs (results significant for each individual population and combined). (E) A survival curve for the total load of deletions >30 kb in Western chimpanzees compared to all other great apes shows a significant excess of deletions in this population. Western chimpanzee populations show the lowest diversity of any of the populations assessed in this study and the most fixed deletions of all chimpanzee species assessed.
