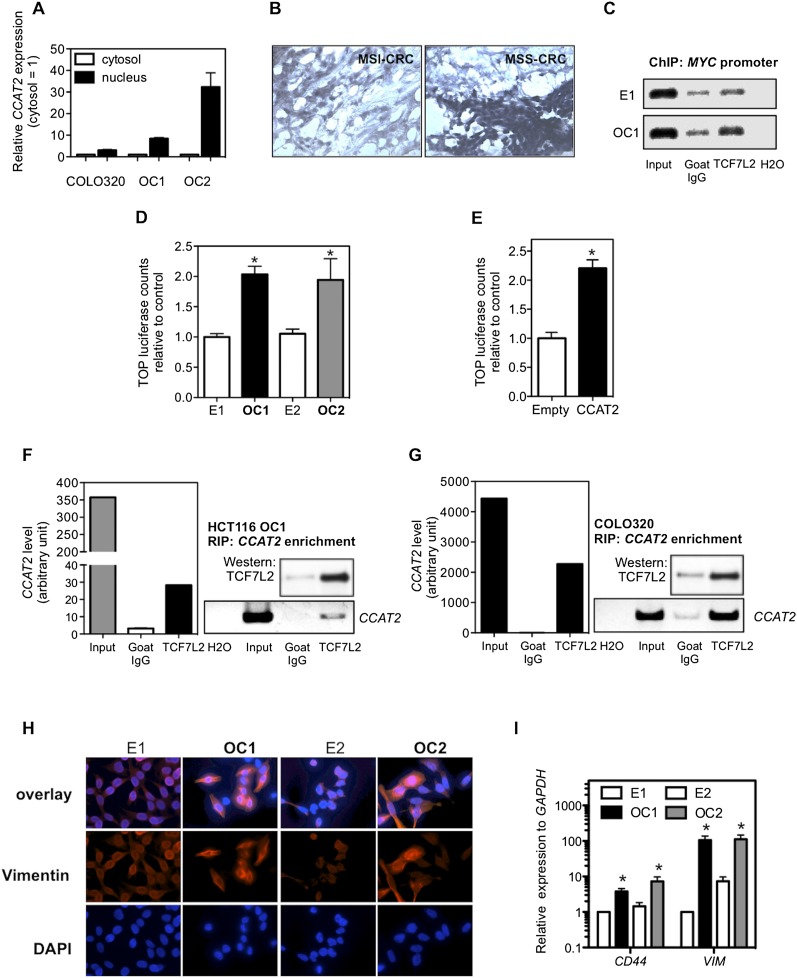
Figure 5.
Regulation of TCF7L2 activity by CCAT2. (A) CCAT2 nuclear localization, as identified using qRT-PCR in fractionated COLO320 cells and HCT116 CCAT2 clones. (B) CCAT2 nuclear localization, as identified in CRC samples using in situ hybridization. (C) CCAT2 increased TCF7L2 binding to the MYC promoter. ChIP analysis showed higher binding in CCAT2-overexpressing clone (OC1) than cells with basal CCAT2 expression (E1). Goat IgG was used as a control antibody. (D) Higher WNT activity in CCAT2 stable clones, as identified by TOP-Flash reporter assay. (E) Transient expression of CCAT2 induced a more than twofold increase of TOP-Flash luciferase activity in HEK293 cells. The data in C and D are presented as mean ± SEM (n = 3). (*) P < 0.05. (F) Colocalization of CCAT2 RNA with TCF7L2 protein by RNA immunoprecipitation using anti-TCF7L2 antibodies in OC1 with ectopic CCAT2 overexpression. (G) Colocalization of CCAT2 RNA with TCF7L2 protein by RNA immunoprecipitation using anti-TCF7L2 in COLO320 cells, which have endogenous high CCAT2 expression. A quantitative analysis using qRT-PCR is shown along with the PCR gel images. (H) Immunostaining of HCT116 clones with vimentin antibody or DAPI. Stronger vimentin signal as seen in epithelial–mesenchymal transition was observed in CCAT2-overexpressing clones than in cells with basal CCAT2 expression. (I) CCAT2 increases the mRNA expression levels of CD44 and VIM in HCT116 clones. The data are presented as mean ± SEM from at least three independent experiments. (*) P < 0.05.