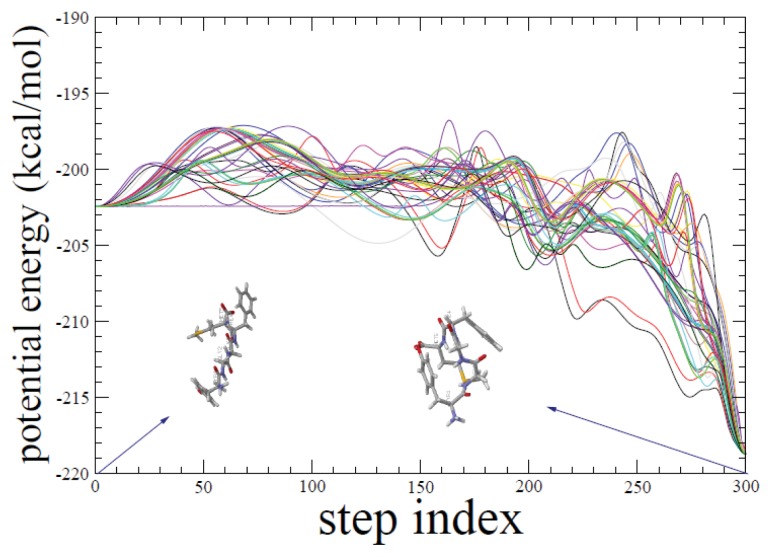
Figure 1.
Fifty independent potential-energy variations for met-enkephalin are shown along the action-derived molecular dynamics (ADMD) step index. The same initial and final conformations are used for all ADMD simulations. The potential-energy variations clearly show the existence of multiple pathways. There exist many alternative folding routes that connect the extended and compact peptide conformations. Two conformations used as inputs for ADMD simulations are shown. The initial (final) conformation shown at j = 0 (300) is prepared by applying local energy minimization to the fully-extended (compact) structure. We observe that one of the ADMD trajectories is of almost constant potential energy in the first half of the trajectory. In this part, the kinetic energy is rather high. It should be noted that the total energy (sum of kinetic energy and potential energy) is conserved along all ADMD pathways.