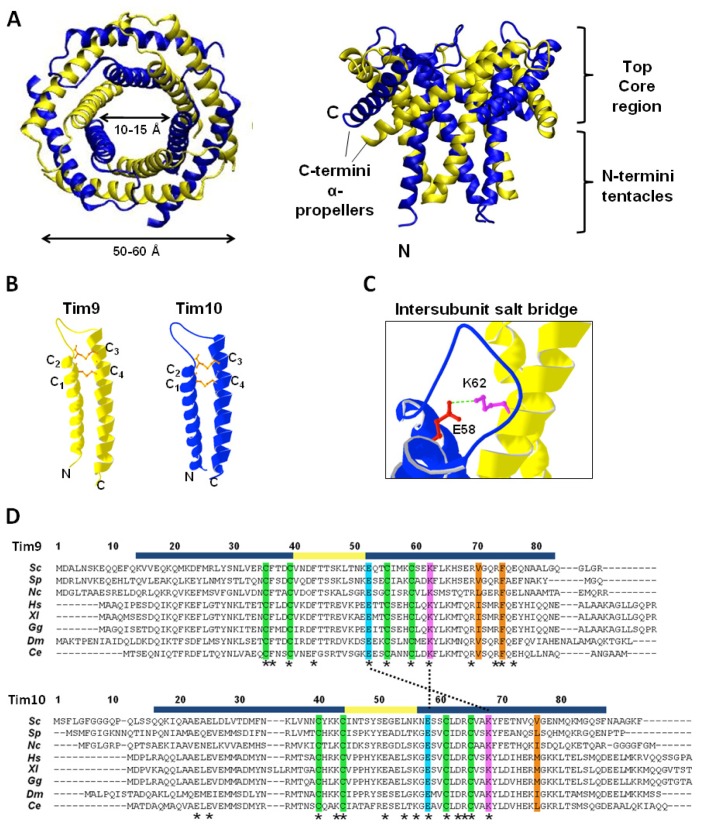
Figure 4.
Tim9–Tim10 hexameric complex. (A) Crystal structure of yeast Tim9–Tim10 complex, top (left) and side (right) view respectively (PDB: 3DXR). Tim9 (yellow) and Tim10 (blue). The outer layer is formed by the six C-terminal helices (α-propellers) and the inner layer by the six N-terminal helices (tentacles); (B) Structure of Tim9 and Tim10 showing the two helices linked by intramolecular disulfide bonds (orange) (PDB: 3DXR); (C) Conserved salt bridge between a glutamate residue in the loop of Tim10 and a lysine residue in the outer helix of Tim9; (D) Alignment of Tim9 and Tim10 amino acid sequences. Fully conserved residues are marked with asterisks (*). Helical regions and the central loop are marked in blue and yellow respectively. Conserved cysteine residues are highlighted in green. Key charged residues are highlighted in pink (positively charged) and light blue (negatively charged), and salt bridges between them are marked with a dotted line. Central residues of hydrophobic clusters are highlighted in orange. Sc: Saccharomyces cerevisiae, Sp: Schizosaccharomyces pombe, Nc: Neurospora crassa, Hs: Homo sapiens, Xl: Xenopus laevis, Gg: Gallus gallus, Dm: Drosophila melanogaster, Ce: Caenorhabditis elegans.