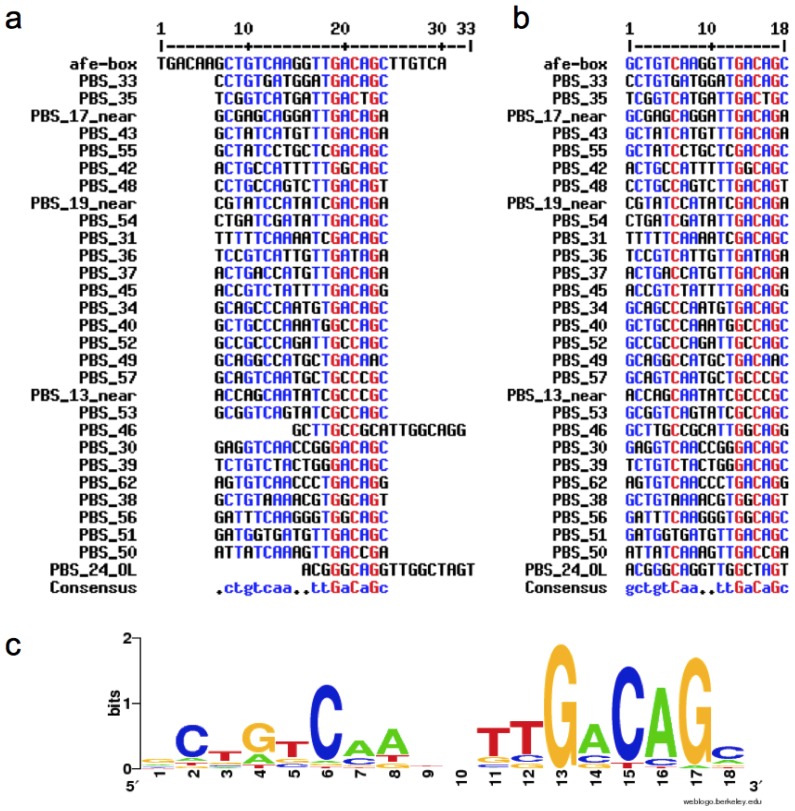
Figure 3.
Multiple sequence alignment of selected predicted binding sites (PBSs) that were also positive for the MEME common motif search. Only MEME motifs located near (“_near”) or overlapping (“_OL”) SP type PBSs are indicated. (a) Alignment including the complete version of the afe-box (30 nucleotides); (b) Alignment including the central palindromic module of the afe-box (18 nucleotides). PBS_24_OL and PBS_46 align with better scoring to the right half of the complete afe-box. Numbers correspond to PBSs listed in Supplementary Table 1; (c) Sequence logo file created from the sequence alignment shown in b.