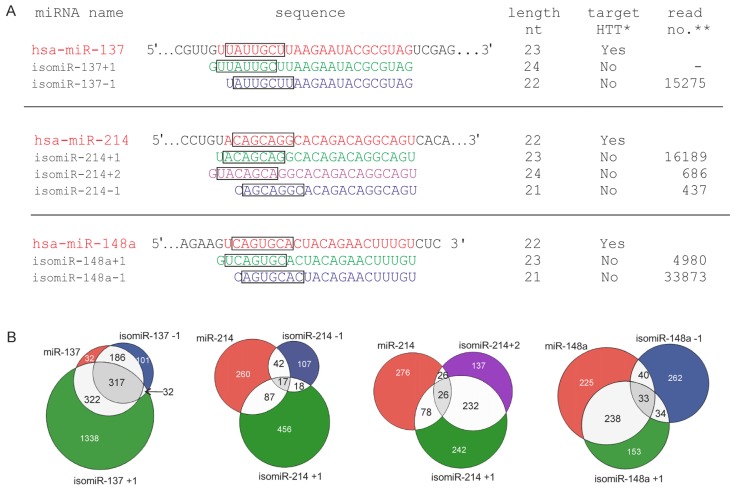
Figure 4.
Graphical presentation of selected isomiR variants and their potential to target different genes. (A) Nucleotide sequences of miR-137, miR-214, miR-148a, and their isoforms. miRNA sequences are marked in red, and isomiR sequences are shown in blue, green, and violet for −1-, +1-, and +2-nt seed shifting, respectively. The miRNA seed sequences are labeled with black rectangles. Information on the miRNA lengths, as well as their potential for targeting the HTT gene and isomiR expression levels, is also provided. (*) Ability to interact with the HTT 3′UTR, as predicted by the TargetScanHuman algorithm (Release 6.2) for miRNAs and the TargetScan Custom (Release 5.2) for isomiRs [46], (**) isomiR read number according to the YM500 database [53]; (B) Venn diagrams showing the predicted miRNA targets for selected isomiRs. Potential targets for the 5′-end variants of miR-137, miR-214, and miR-148a were predicted using the TargetScan Custom algorithm (Release 5.2) and are shown as overlaps in the Venn diagrams. Targets for the canonical miRNAs are compared with the targets for the miRNAs with the shifted seed regions and are depicted in the same colors as in panel A. The numbers inside the circles denote the numbers of potential targets predicted for the appropriate miRNA variants.