Figure 2. Individual class I HLA binding of peptides derived from mutated BCR-ABL in CML patients.
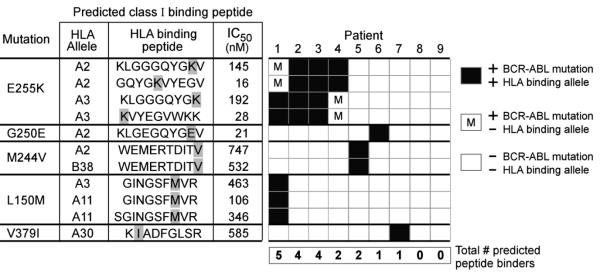
Eight distinct BCR-ABL mutations were detected among 9 patients with imatinib resistant CML. Displayed are the 5 of 8 mutations for which HLA binding peptides could be predicted from the individual patients, and are ordered by decreasing frequency based on larger patient series (37). For each mutation, class I HLA binding peptides using only the HLA alleles represented within the set of 9 patients that are predicted by the NetMHC algorithm are provided. Not shown are the three mutations that were present in patient leukemia cells, but based on the patients’ HLA typing, were not predicted to generate any HLA-binding peptides (T315I [for Pts 1, 4, 8); E281K [for Pts 5,9], and T315A [Pt 5]). Black boxes - patients expressing both the HLA allele and the BCR-ABL mutation that would generate a binding peptide with affinity of IC50<1000 nM. White boxes - patients without expression of a particular BCR-ABL mutation. White box with “m”-patients harboring a particular BCR-ABL mutation, but not an HLA allele that would be predicted to bind the mutated peptide.
