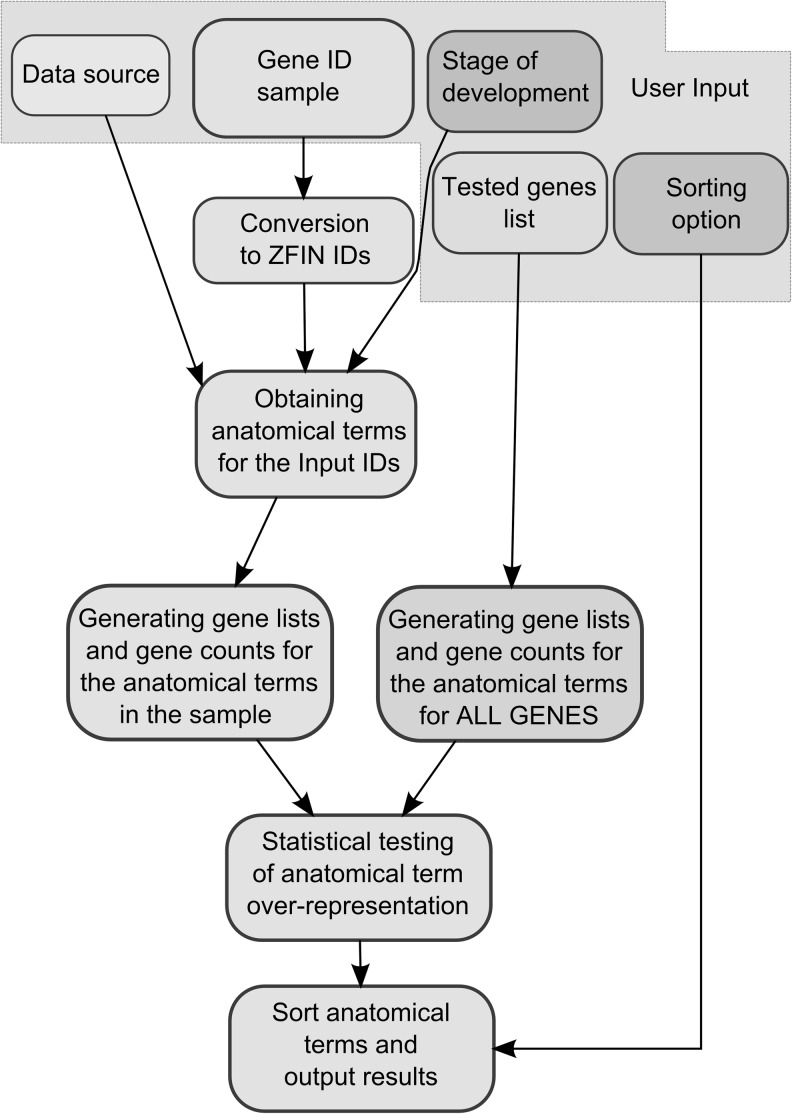
FIG. 2.
Scheme of the overall ZEOGS software algorithm. To run ZEOGS, the user chooses a data source, a gene identifier list (either gene symbol or ZFIN ID), stage of development for which the output will be generated, a list of tested genes, and a sorting option for the retrieved anatomical terms. Input items are surrounded by a gray area. Next, the input gene identifiers are converted to ZFIN IDs to query the gene expression datasets according to the user-selected data source and the stage of development. The output consists of a list of anatomical terms assigned to genes in the sample, followed by a list of genes and their associated counts for all retrieved anatomical terms. The above procedure can be performed either for all the zebrafish genes with available expression information or for the list of genes actually tested in an experiment. Enrichment of anatomical terms is assessed by means of the p-value from a hypergeometric test. Finally, anatomical terms are sorted according to the user-selected sorting option.