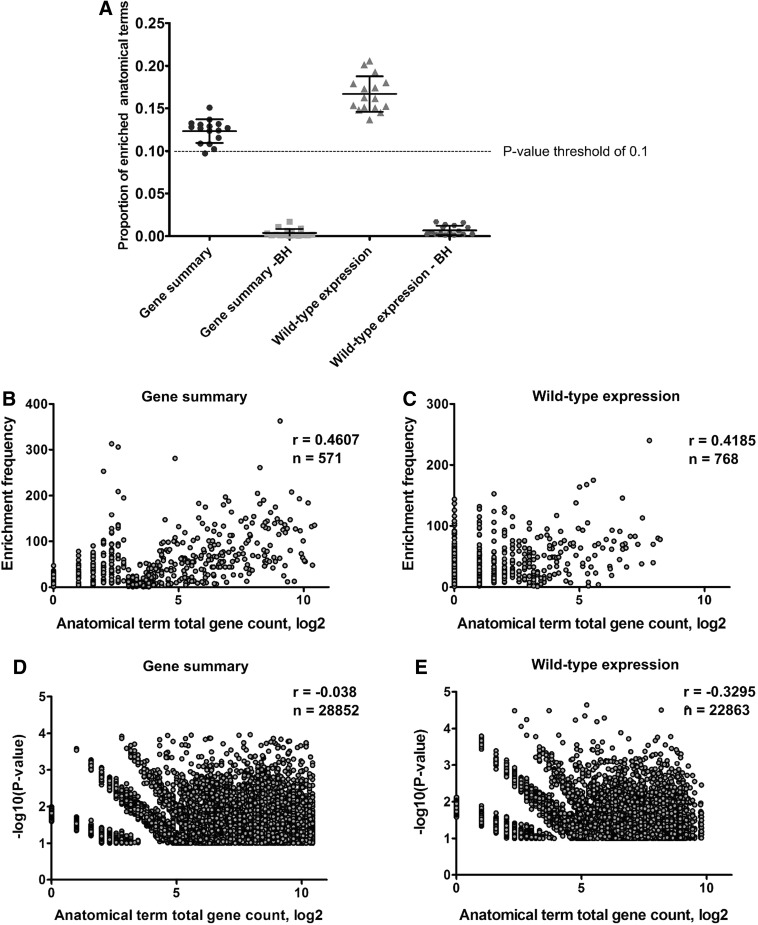
FIG. 4.
Random sampling experiment using ZEOGS to assess the false-discovery rate and the possible bias toward high gene counts. A basic ZEOGS algorithm was run 200 times at stages 17–33 (5–60 hpf) for both “gene summary” and “wild-type expression” data source options using a random selection of 500 genes. The proportion of anatomical terms for which the enrichment p-value was below 0.1 (enrichment threshold) was recorded as well as the number of associated genes. Data points represent fractions of enriched anatomical terms at individual stages for both data source options and a 0.1 p-value cutoff are presented when standard hypergeometric p-value calculations or the Benjamini–Hochberg-corrected p-values (BH) were done (A). Plots of the number of times each anatomical term was found enriched (enrichment frequency) relative to the log2 of the anatomical term's total gene count for all stages combined show a modest correlation between these two variables (Spearman coefficient r) for both “gene summary” (B) and “wild-type expression” (C) data source options. The extent of enrichment measured by the negative log10 of p-values did not correlate with log2 of the total gene counts of anatomical terms for either of “gene summary” (D) or “wild-type expression” (E) data source options.