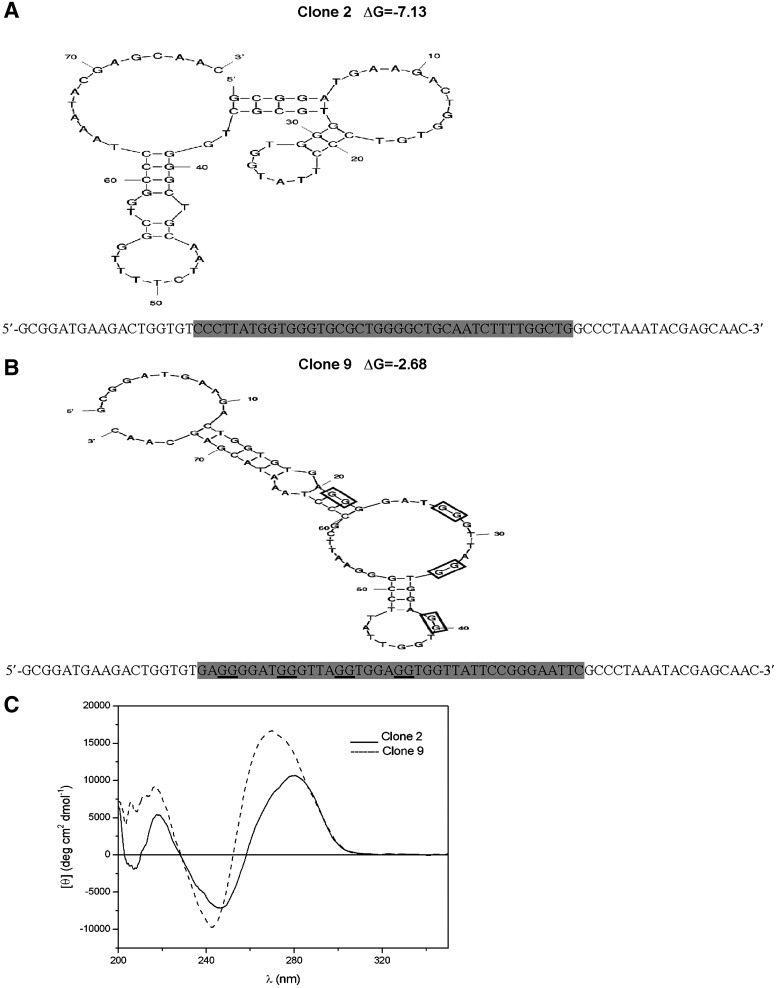
FIG. 3.
Sequence and secondary structure of ABA-binding subclones from the SEL7 population. Predicted structures of clone 2 (A) and clone 9 (B). Structures and free energy values (ΔG, in kcal/mol) were obtained using mfold as described in Materials and Methods. The 40-nucleotide variable region of each aptamer (highlighted in gray) is flanked by two 18-nt constant primer binding regions. The underlined G-doublets in clone number 9 are those with the highest probability (G-score) of participating to the G-quadruplex formation based on the algorithm used by the QGRS Mapper software. (C) The single-stranded (ss) DNA aptamers (10 μM) were thermally denatured and then cooled to 20°C before obtaining the circular dichroism (CD) spectra: shown is the average of three scans from 200 to 350 nm at a rate of 20 nm per minute with a step resolution of 0.2 nm, a time constant of 2 seconds, and a bandwidth of 2.0 nm. Spectra were corrected by subtracting the appropriate buffer background and normalized to mean residue ellipticity.