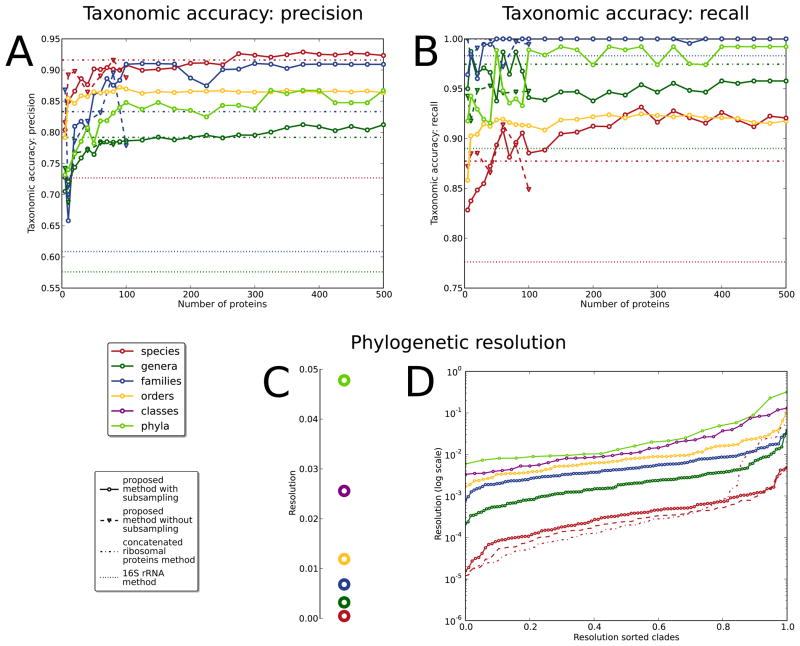
Figure 2. Selecting informative subsequences improves the accuracy of phylogenetic tree reconstruction.
As compared to a gold standard derived from the IMG taxonomy, both precision (A) and recall (B) of inferred phylogenies increase at all taxonomic levels as up to the 500 most-conserved proteins are sampled (values averaged across all clades at each level). Comparison with full-length protein sequence phylogenies (up to 100 proteins) confirms that alignments subsampled at the most discriminative amino acids are both more accurate and more efficient. This approach outperforms single 16S rRNA gene phylogenies at all taxonomic levels, as well as trees based on curated ribosomal protein concatenation15,16 for all but the most specific clades. (C) The relative phylogenetic diversity of all taxonomic levels is consistent across varying protein numbers and is on average remarkably logarithmic, providing quantitative support for the existing multi-level microbial taxonomy. (D) Relative phylogenetic diversity among individual clades at each taxonomic level, however, shows a tremendous range of diversities, with some underrepresented phyla comprising only as much sequence divergence among available genomes as some species. This suggests that while taxonomic levels are consistent on average, clade-specific diversity thresholds should be employed when linking phylogenetic divergence with individual taxonomic labels. Again, even the most diverse species reconstructed by this method are better resolved than those using the 16S rRNA gene alone, for which many demonstrate improbably high putative phylogenetic diversity.