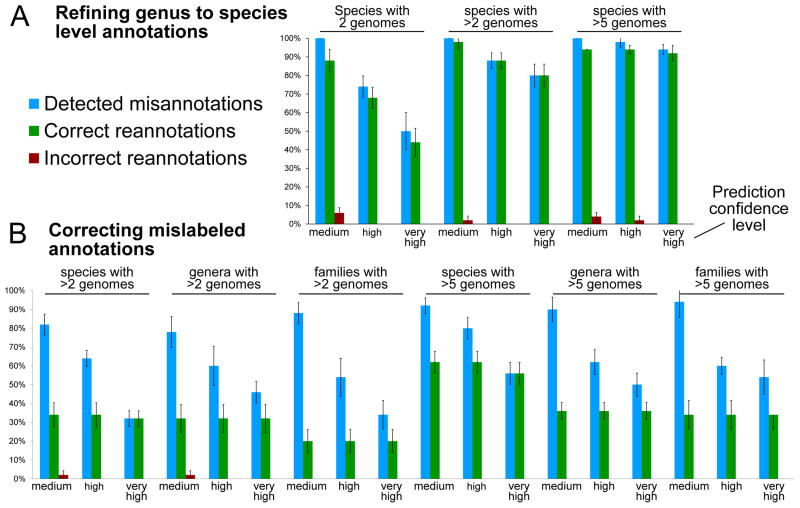
Figure 4. Accuracy of correctly re-inferred taxonomic labels for artificially-mislabeled organisms.
Barplots report the percentages (with s.d.) of successfully-recovered cases. (A) For 5 iterations, 10 taxa are selected at random from species with 2, more than 2, or more than 5 genomes, and their species-level label removed. The PhyloPhlAn phylogenetic tree (which is built without any taxonomic information) is then used to re-impute the removed labels at medium, high, and very high confidence thresholds. No incorrect refinements are produced at the highest confidence threshold, and average recall rates for species with at least three taxa exceed 90% at high confidence. (B) We repeat this procedure by mislabeling (rather than removing labels for) species, genus, or family-level assignments. No false positives are produced at high or very high confidence, and only 2 over all experiments (<1%).