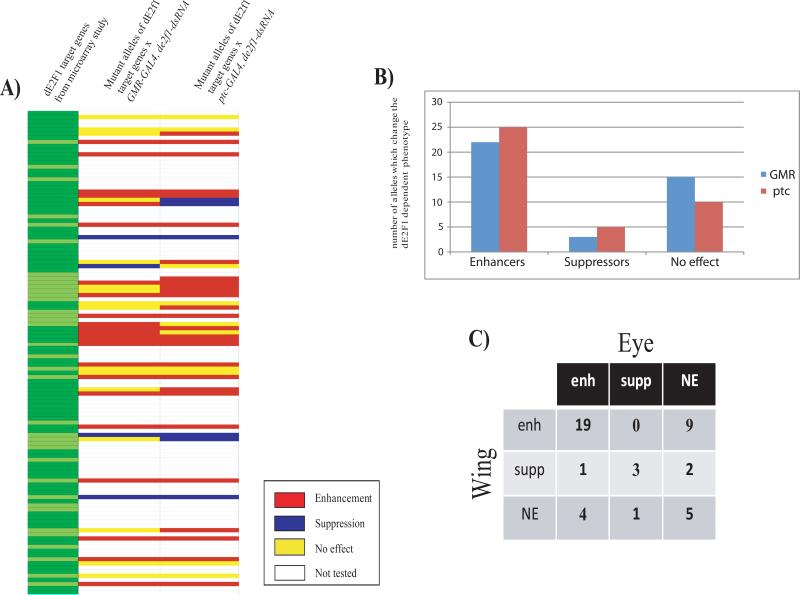
FIGURE 1. Mutant alleles of many dE2F1-dependent genes modify dE2F1-dependent phenotypes.
1A) A heat map summarizing the genetic interactions observed when mutant alleles of dE2F1-regulated genes were tested for their ability to dominantly modify visible phenotypes caused by dsRNA-mediated targeting of dE2F1 in the eye (GMR-GAL4;de2f1-dsRNA) or the wing (ptc-GAL4;de2f1-dsRNA). Alleles were tested for genes of dE2F1 targets that were identified in previous microarray studies (Dimova et al., 2003). Enhancers are shown in red, suppressors in blue, mutant alleles that had no effect are shown in yellow and alleles which where not tested in white. For details of the alleles used and the strength of the interaction see Table 1.
1B) Bar graph illustrating the number of mutant alleles that enhanced, suppressed, or had no effect on the eye phenotype (GMR, blue) or the wing phenotype (ptc, red).
1C) Table showing the actual number of mutants which either affected both eye and wing phenotype, which affected one but not the other, or which had no effect on either of the two phenotypes. Abbreviations: enhanced (enh), suppressed (supp) or no effect (NE).