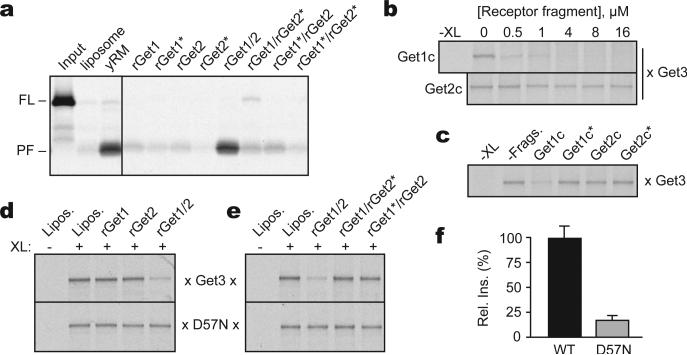
Figure 4. Mutational analysis of Get1/2/3 function.
(a) Insertion assay with purified Get3-Sec61β targeting complex and proteoliposomes containing the indicated purified proteins. Liposomes and yRM are controls. Get1* and Get2* indicate mutants inactive in Get3 interaction (R73E and R17E, respectively). (b) Substrate-release from targeting complexes incubated with Get1c or Get2c; release was monitored by loss of the crosslink between radiolabeled substrate and Get3. (c) As in (b), with wild-type and mutant fragments at 0.5 μM. (d) Substrate interaction with Get3 or the ATPase-deficient Get3(D57N) was assessed by crosslinking after incubation with liposomes or proteoliposomes containing the indicated recombinant proteins. (e) As in (d), but comparing wild-type and mutant Get1/2 complexes. (f) Relative insertion efficiency into rGet1/2 proteoliposomes of targeting complexes prepared with wild-type Get3 or Get3(D57N).