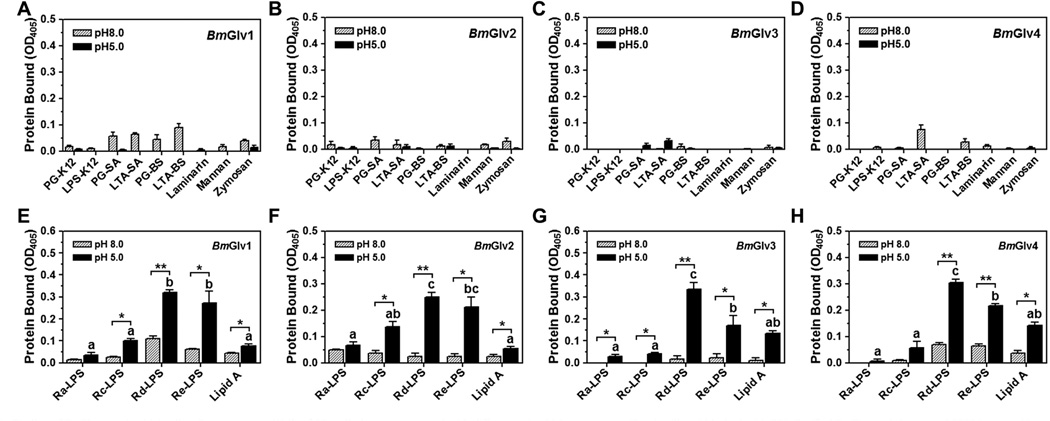
Fig. 3.
Binding of BmGlvs to microbial cell wall components. Wells of 96-well plates were coated with different microbial components (2 µg/well) and blocked with BSA. Purified BmGlvs and the control CP36 (a recombinant cuticle protein from M. sexta) were diluted to 1 µg/ml in 10 mM phosphate, 100 mM NaCl, pH 5.0 or 8.0, and the diluted proteins were added to the coated plates. Binding of proteins to microbial components was determined by plate ELISA assays as described in the materials and methods. The figure showed specific binding of BmGlvs to each microbial component after subtracting the total binding of the control CP36 protein from the total binding of BmGlvs. Each bar represents the mean of at least three individual measurements ± SEM. Comparing binding of each BmGlv to rough LPS and lipid A at pH 5.0 (panels E–H), identical letters are not significant difference (p > 0.05), while different letters indicate significant difference ( < 0.05) determined by one way ANOVA followed by a Tukey’s multiple comparison test. Comparing binding of BmGlv to each rough LPS or lipid A between pH 5.0 and 8.0 (panels E–H), ‘*’ (p < 0.05) and ‘**’ (p < 0.01) indicate significant differences determined by an unpaired t-test