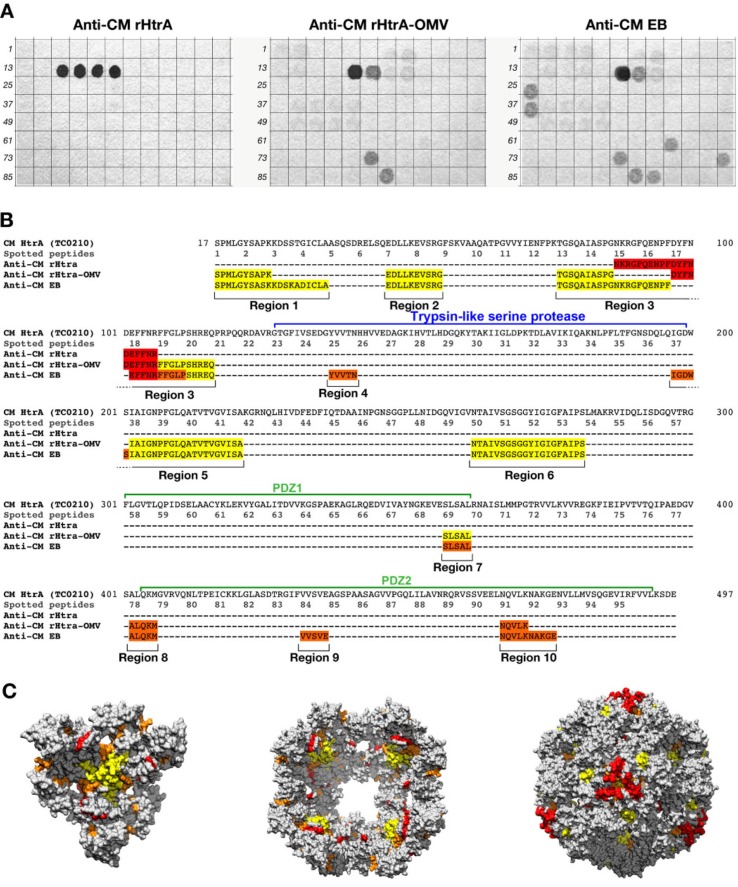
Fig. 5.
Differential epitope recognition pattern of CM rHtrA and CM rHtrA-OMV antisera. (A) Peptide arrays (96 peptides) were prepared on a cellulose membrane by in situ synthesis of overlapping sequences of 15-residue peptides covering the amino acid sequence of the mature CM HtrA form and tested for binding by immunoblotting with polyclonal antibodies directed against CM rHtrA-OMV (left), CM rHtrA-OMV (centre) and CM EBs (right). Membranes incubated with serum raised against empty OMVs and run simultaneously gave negative staining. Numbers on the left of each blot mark the first peptide of each row of the arrays. (B) Colour code representation of the amino acid regions recognized by the different antisera on the mature from of the CM HtrA protein sequence. The sequences recognized by each antiserum are highlighted according to the staining intensity of spotted peptides: red, high intensity; orange, moderate intensity; yellow, low intensity. (C) Molecular modelling of CM HtrA multimeric forms. The 3-mer basic structure that assembles in multiple copies to form both the bowl-shaped, lipid-associated structures and the spherical soluble structures is shown on the left. The 4-fold symmetry bowl-like structure proposed by Shen and colleagues (29) is reported in the central panel of Fig. 5C looking at the solvent-exposed concave face of the structure. Finally, the spherical structure of 24-mer soluble HtrA is reported in the right-hand side of the figure. The regions recognized by anti-CM rHtrA and anti-CM rHtrA-OMV antibodies are highlighted in the 3 models maintaining the same colour code as in (B). The models were generated from PDB structure 3cs0.