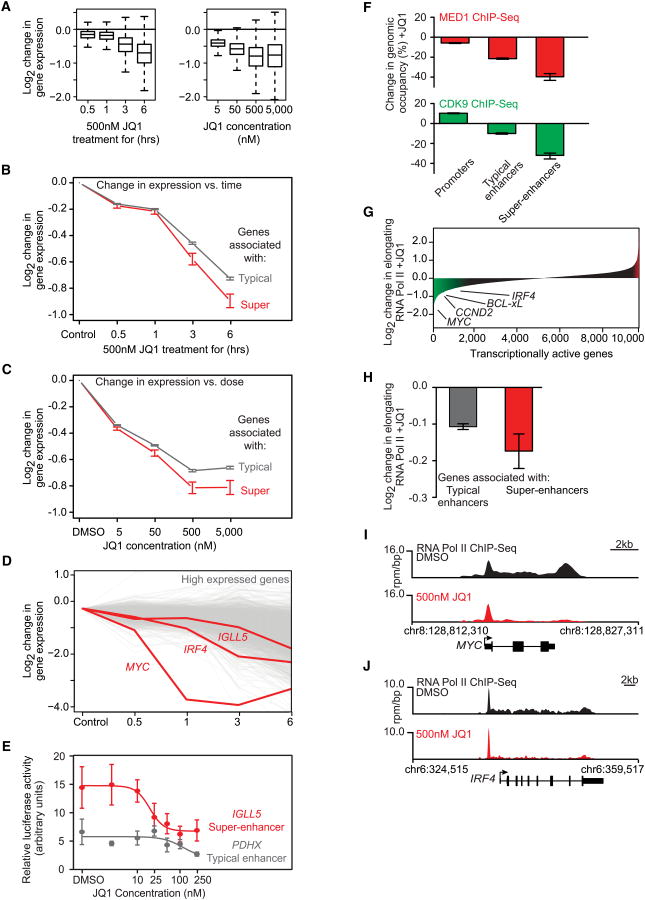
Figure 6. JQ1 Causes Disproportionate Loss of Transcription at Super-Enhancer Genes.
(A) Box plots showing the Log2 change in gene expression for all actively transcribed genes in JQ1-treated versus control cells for a time course of cells treated with 500 nM JQ1 (left) or for a concentration course of cells treated for 6 hr with varying amounts of JQ1 (right). The y axis shows the Log2 change in gene expression versus untreated control cells (left graph) or control cells treated with DMSO for 6 hr (right graph).
(B and C) Line graph showing the Log2 change in gene expression versus control cells after JQ1 treatment in a time- (B) or dose (C)-dependent manner for genes associated with typical enhancers (gray line) or genes associated with super-enhancers (red line). The y axis shows the Log2 change in gene expression of JQ1 treated versus untreated control cells. The x axis shows time of 500 nM JQ1 treatment (B) or JQ1 treatment concentration at 6 hr (C). Error bars represent 95% confidence intervals of the mean (95% CI).
(D) Graph showing the Log2 change in gene expression after JQ1 treatment over time for genes ranked in the top 10% of expression in MM1.S cells. Each line represents a single gene, with the MYC and IRF4 genes drawn in red. The y axis shows the Log2 change in gene expression of JQ1-treated versus untreated control cells. The x axis shows time of 500 nM JQ1 treatment.
(E) Line graph showing luciferase activity after JQ1 treatment at various concentrations for luciferase reporter constructs containing either a fragment from the IGLL5 super-enhancer (red line) or the PDHX typical enhancer (gray line). The y axis represents relative luciferase activity in arbitrary units. The x axis shows JQ1 concentrations. Error bars are SEM.
(F) Bar graphs showing the percentage loss of either MED1 (top, red) or CDK9 (bottom, green) at promoters, typical enhancers, and super-enhancers. Error bars represent 95% CI.
(G) Graph of loss of RNA Pol II density in the elongating gene body region for all transcriptionally active genes in MM1.S cells after 6 hr of 500 nM JQ1 treatment. Genes are ordered by decrease in elongating RNA Pol II in units of Log2 fold loss. Genes with a greater than 0.5 Log2 fold change in elongating RNA Pol II are shaded in green (loss) or red (gain). The amount of RNA Pol II loss is indicated for select genes.
(H) Bar graph showing the Log2 fold change in RNA Pol II density in elongating gene body regions after 6 hr of 500 nM JQ1 treatment for genes with typical enhancers (left, gray) or genes with super-enhancers (red, right). Error bars represent 95% confidence intervals of the mean (95% CI).
(I and J) Gene tracks of RNA Pol II ChIP-seq occupancy after DMSO (black) or 500 nM JQ1 treatment (red) at the super-enhancer proximal MYC gene (I) and IRF4 gene (J). The y axis shows signal of ChIP-seq occupancy in units of rpm/bp.
See also Figure S3.