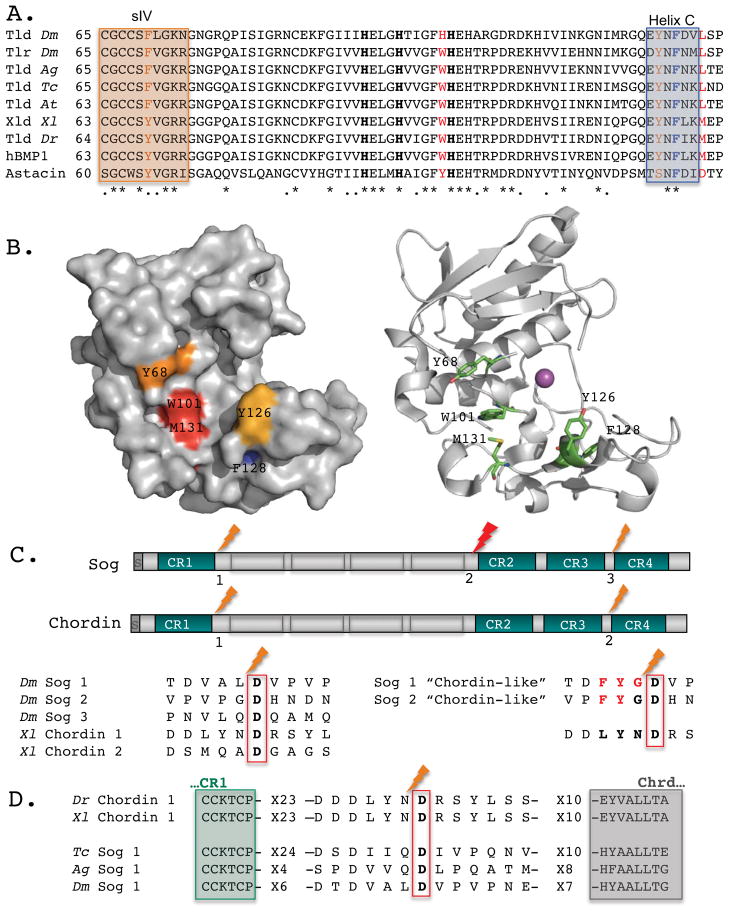
Figure 2. Tlds catalytic domains and their processing sites in Sog/Chordin.
(A) ClustalW alignment reveals highly conserved aromatic residues within the catalytic domain of Tld-type enzymes in Drosophila melanogaster (Dm), Anopheles gambiae (Ag), Tribolium castaneum (Tc), Achaearanea tepidariorum (At), Xenopus laevis (Xl), Danio rerio (Dr), and humans (hBMP-1) as compared with the crayfish astacin. Three zinc-binding His residues are shown in bold. Crystal structure of hBMP-1 catalytic domain indicates that P2 residue of the substrate extends towards Trp101 and Met131, shown in red [95]. Boxes mark the β-sheet strand (sIV) on the upper side of the catalytic pocket and Helix C on the lower side. These motifs contain additional aromatic residues that may come in close proximity to P3 (brown). Helix C also includes F128 (blue), a residue mutated in autosomal recessive osteogenesis imperfecta [96]. (B) A surface diagram of hBMP-1 catalytic domain (left) and a ribbon diagram (right) capture the cavity where P3 and P2 residues bind. The conserved residues discussed are color coded (left) or shown as sticks colored by elements (right) (C, green; O, red; N, blue). The catalytic zinc is shown as a magenta sphere. (C) Drosophila Sog and Xenopus Chordin shared similar organization, with four Cysteine-rich von Willenbrandt factor C domains (CR) separated by four “Chordin-like” (Chrd) motifs. The Tld-processing sites in Sog and Chordin show little conservation besides the S1′ Asp residue, a hallmark of this family of protease. Several residues (P1–P3) are responsible for making Sog dependent on BMP for Tld processing, while Chordin is not. Changes at these positions (shown in red) make Sog a BMP-independent substrate for Tld, “Chordin-like” [49]. (D) Comparison of Tld processing sites, residing between CR1 and Chrd repeats, suggests that Tribolium and Anopheles Sog are BMP-dependent for their processing, resembling the Drosophila Sog.