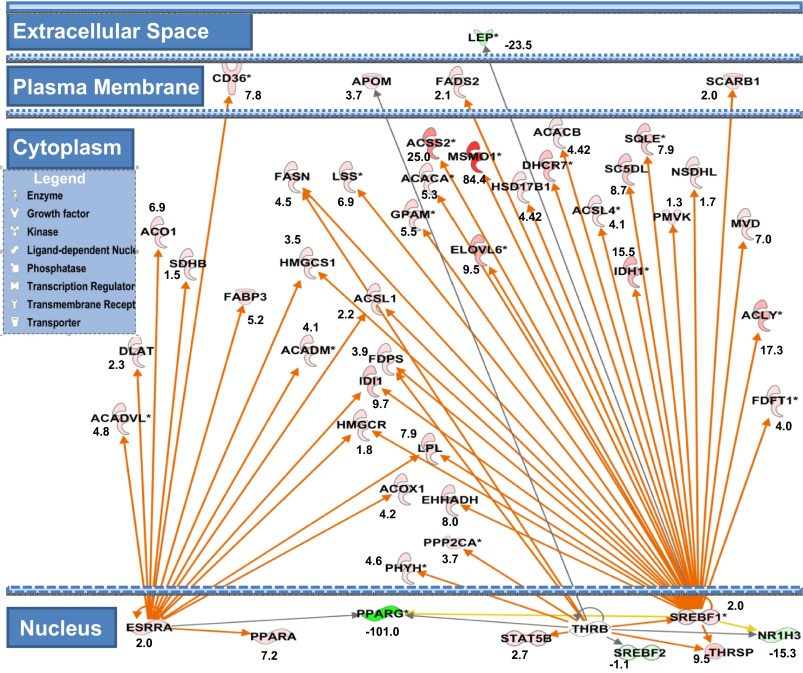
Fig. 7.
Cell compartment view for network of lipid metabolic genes highlighting the role of SREBF1, THRSP, and ESRRA curated by ingenuity pathway analysis (IPA). Green nodes indicate downregulated genes; red indicates upregulated genes, and numbers represent fold changes at 72 h vs. 6 h. Orange arrows indicate predicted activation, yellow indicate inconsistent findings with state of the downstream gene, and gray indicate that the relationship is not predicted. Activation of SREBF1 leads to increase in expression of the genes for lipases (LPL), HDL transporter (SCARB1), TCA cycle (IDH), de novo FA synthesis (ACACA&B, ACLY, FASN), FA activation (ACSS1&2, ACSL1&4), FA elongation (ELVOL6), FA desaturation (FADS2, SC5DL), TG synthesis (GPAM), whereas the remaining genes are primarily involved in sterol biosynthesis. Downstream of ESRRA activation is the induction of the transcription factor (PPARA), FA transporter (CD36), lipase (LPL), FA binding (FABP3), TCA (SDHB, ACO1), FA modification (ACOX1, ACADM, ACADVL), FA activation (ACSL1), and others for cholesterol synthesis (HMGCS1, HMGCR, DLAT). Activation of THRB upregulates transcription factor (STAT5B) and de novo FA synthesis regulator (THRSP) as well as FA synthesis (FASN), FA activation (ACSL1), FA oxidation (PHYH, EHHADH), protein phosphatase (PPP2CA), and sterol synthesis (HMGCR, IDI1).