Abstract
Large-scale computational analyses of the growing wealth of genome-variation data consistently tell two distinct stories. The first is expected: coding variants reported in disease-related databases significantly alter the function of affected proteins. The second is surprising: the genomes of healthy individuals appear to carry many variants that are predicted to have some effect on function. As long as the complete experimental analysis of all human genome variants remains impossible, computational methods, such as PolyPhen, SNAP, and SIFT, might provide important insights. These methods capture the effects of particular variants very well and can highlight trends in populations of variants. Diseases are, arguably, extreme phenotypic variations and are often attributable to one or a few severely functionally disruptive variants. Our findings suggest a genomic basis of the different nondisease phenotypes. Prediction methods indicate that variants in seemingly healthy individuals tend to be neutral or weakly disruptive for protein molecular function. These variant effects are predicted to be largely either experimentally undetectable or are not deemed significant enough to be published. This may suggest that nondisease phenotypes arise through combinations of many variants whose effects are weakly nonneutral (damaging or enhancing) to the molecular protein function but fall within the wild-type range of overall physiological function.
Keywords: nsSNP, coding SNV, variome analysis, genomic variant burden, evolution
Scientists have been exploring the genetic basis of disease ever since the first forays into genome sequencing (1) and before (2). Nonsynonymous single-nucleotide polymorphisms (nsSNPs) (here, nonsynonymous single-nucleotide variants of any population frequency), variants that alter the amino acid sequence, have been associated with many diseases (3). Because experimental findings are rarely reported to databases in a standardized form, metaresources, such as the Protein Mutant Database (PMD) (4) and Swiss-Prot/UniProt (5), manually curate publications to map variants to diseases. Natural language processing automates this mapping (6).
However, the experimental data are inherently biased for variants affecting function (nonneutral; enhancing or damaging protein function) over those that do not (neutral; no functional difference between wild type and mutant). First, scientifically, it is more interesting to study variants that have an effect than those that do not. Second, proving that a mutation has an effect requires only one successful experiment. Proving that a mutation is truly neutral requires ascertaining neutrality for all possible functional assays. In fact, we are not aware of any experimental study to date that has conducted such a comprehensive exploration of neutrality for any significant set of variants. Computational predictions, although more accurate than some high-throughput experiments (7), are never as specific as careful, detailed experiments. Importantly, however, predictions do not have the neutrality problem; computational evaluation of variant effects (8, 9) is inexpensive and fast, allowing rapid testing of all possible point mutations without preselection.
At least one study suggests that healthy people carry hundreds of severely functionally damaging mutations, some of which are disease-associated (10). If so, their genomes may also contain many weakly damaging (nonneutral) variants (11), as predictions also indicate. Do computational methods overpredict functional changes, or is the “neutral range” of variant effects wider than we expect? Enough sequencing data are now available to consider these questions in detail.
We used our computational method screening for nonacceptable polymorphisms (SNAP) (8) to trace patterns in available variant sets (Table 1 and SI Methods). SNAP uses machine learning (neural network) to predict functional effects of amino acid variants on the basis of protein sequence features, such as conservation, predicted secondary structure, and solvent accessibility. Although other methods, such as sorting intolerant from tolerant (SIFT) (12) and polymorphism phenotyping (PolyPhen) (9), address similar objectives, they either do not exhibit the same ability to stratify predictions by effect severity and/or focus on different goals. Nevertheless, we also investigated results from SIFT and PolyPhen-2. We find that (i) known disease-causing variants are almost always predicted to have severe impact on function and (ii) most variants in the genomes of healthy individuals are likely to be experimentally classified as neutral (identical to wild type) given current state-of-the-art laboratory techniques. However, we also predict that (iii) nearly half of the variants from healthy individuals render the affected genes/proteins functionally different from the “reference” genome “wild type.” This suggests a fairly wide range of physiologically acceptable levels of protein function that is spanned by human diversity. Individuals might then differ from one another via many functionally minor changes within this “physiologically wild-type range.”
Table 1.
Functional annotations of variant datasets
| Set | Total | Subset | Contents | Exp. Pos | Exp. Neg | SNAP Pos | SNAP Neg | Total in subset |
| EC | 26,787 | # | Single amino acid differences in enzymes of same function | 0 | 26,787 | 1,344 | 25,443 | 26,787 |
| Syn | 9,228 | # | Variants of form XposX in all positions (pos) of PMDneutral | 0 | 9,228 | 423 | 8,805 | 9,228 |
| PMD | 5,5704 | Neut | Experimentally annotated “no” effect on protein function | 0 | 14,651 | 7,366 | 7,285 | 14,651 |
| Mild | Experimentally annotated “mild” functional effect | 20,884 | 0 | 14,346 | 6,538 | 20,884 | ||
| Mod | Experimentally annotated “moderate” functional effect | 4,393 | 0 | 3,501 | 892 | 4,393 | ||
| Sev | Experimentally annotated “severe” functional effect | 15,776 | 0 | 13,582 | 2,194 | 15,776 | ||
| PMD/EC-Human | 20,143 | EC | Subset of the ECs affecting human proteins | 0 | 4,105 | 232 | 3,873 | 4,105 |
| Neut | Subset of the PMDneutrals affecting human proteins | 0 | 3,271 | 1,623 | 1,648 | 3,271 | ||
| Mild | Subset of the PMDmilds affecting human proteins | 4,886 | 0 | 3,353 | 1,533 | 4,886 | ||
| Mod | Subset of the PMDmoderates affecting human proteins | 933 | 0 | 721 | 212 | 933 | ||
| Sev | Subset of the PMDseveres affecting human proteins | 6,948 | 0 | 5,674 | 1,274 | 6,948 | ||
| PMDstr | 8,326 | # | Experimentally annotated structure/stability effect | 4,087+* | 1,139+* | 5,557 | 2,769 | 8,326 |
| L&L | 6,056 | Neut | Experimentally annotated L&L “no” functional effect | 0 | 3,644 | 1,211 | 2,433 | 3,644 |
| Inter | Experimentally annotated L&L “intermediate” functional effect | 1,071 | 0 | 680 | 391 | 1,071 | ||
| Sev | Experimentally annotated L&L “severe” functional effect | 1,341 | 0 | 1,100 | 241 | 1,341 | ||
| SWISSPROT | 65,654 | Dis | Annotated by Swiss-Prot as disease variants | * | * | 23,337 | 3,672 | 27,009 |
| Poly | Annotated by Swiss-Prot as polymorphisms | * | * | 23,395 | 15,250 | 38,645 | ||
| 1KG | 384,062 | # | Variants from the 1000 Genomes Project | * | * | 171,066 | 212,996 | 384,062 |
| HapMap | 4,076 | # | Variants from HapMap present with MAF > 0.1 in all populations | * | * | 1,873 | 2,203 | 4,076 |
| FullProt | 498,706 | Pos | All SNP-possible single amino acid changes in 100 human proteins | * | * | 113,810 | 98,392 | 212,202 |
| Imp | All multi-NP single amino acid changes in 100 human proteins | * | * | 185,710 | 100,794 | 286,504 |
Dataset details are in SI Methods. In column headers: Exp, experimental functional effects; Neg, neutral variants; Pos, nonneutrals; SNAP, computational predictions. In rows: subset names are shortened; # indicates the whole set.
Variants for which no functional annotation is available.
Results
Predictions Accurately Distinguish True Neutrals from Nonneutrals.
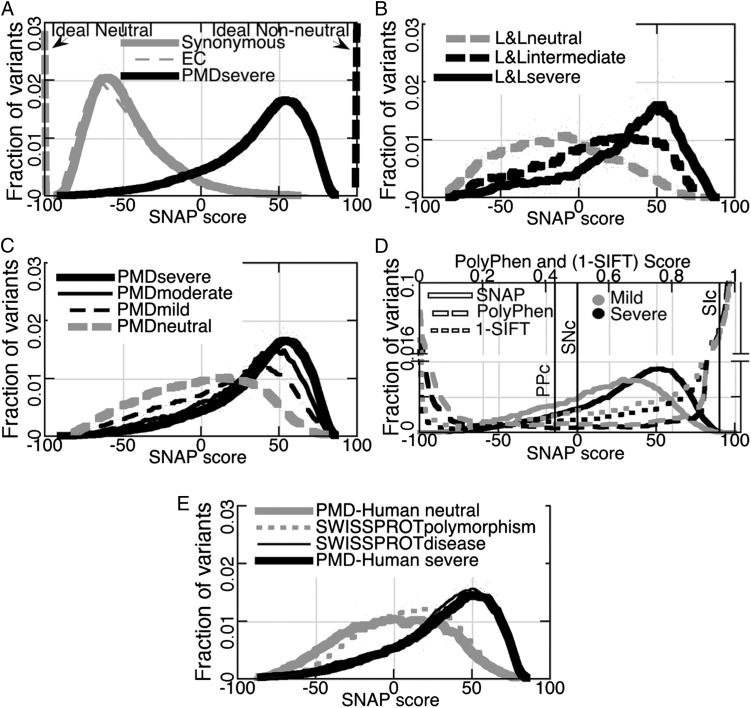
We extracted variants with experimentally established functional effects from PMD (SI Methods and Table 1) and applied SNAP to predict their functional effects. First, we established SNAP’s accuracy limits for large datasets: not all variants with severe effects are predicted with highest nonneutrality scores (+100; Fig. 1A, “PMDsevere”; and Fig. 2, mean/median: 37/44) and not all synonymous variants (Fig. 1A, “Synonymous”; Fig. 2, mean/median: −47/−53) or variants between enzymes with identical function [defined by Enzyme Commission (“EC”) numbers; Fig. 1A, EC; and Fig. 2, mean/median: −49/−55] are predicted with the highest neutrality scores (−100). Because both SNAP predictions and experimental annotations contain some error, it is expected that these distributions do not exactly follow the theoretical ideals (dashed lines at −100/+100 in Fig. 1A). However, the distributions of effect and neutral predictions were significantly different from each other (Tables S1 and S2). These results might slightly over-estimate the power of SNAP because PMDsevere and EC had been used for SNAP training [i.e., SNAP might predict these variants better (closer to −100 for EC and closer to +100 for PMDsevere) than if it had never “seen” them before]. However, SNAP performance differed only marginally between training and testing (8). Hence, we do not expect this effect to be large. SNAP had never been trained to recognize synonymous variants as neutral. The Synonymous distribution in Fig. 1A (overlaying the ECs) is, therefore, conservative.
Fig. 1.
Distribution of variant functional effects. In A–E, the right side of the graph is nonneutral and the left side is neutral for all scoring functions. Curves were generated using SynergySoftware KaleidaGraph v.4.1.3 smoothing via Stineman function (32). (A) Ideally, neutral substitutions should score −100 (vertical gray dashed line) and nonneutrals +100 (vertical black dashed line). Neutral distributions are represented by the Synonymous (gray line) and the EC sets (thin gray dashes). Nonneutrals are the experimental severe functional effect variants (PMDsevere, black line). (B) The L&L variants of severe effect (black line) score higher than intermediates (black dashes). Both sets differ from neutrals (gray dashes). (C) PMDseveres (thick black line) are more right shifted than moderates (thin black line), milds (black dots), or neutrals (gray line). (D) Differentiating the severity of functional effects (PMD/EC-Human set effects: gray, mild; black, severe) using PolyPhen-2 (dashed lines) or SIFT (dotted lines) is impossible. However, higher SNAP scores (solid lines) mean more severe effects. Default binary prediction cutoffs (black vertical lines) are PPc, SNc, and SIc (PolyPhen-2, SNAP, and SIFT, respectively). (E) SWISSPROTdisease variants (thin black line) are as nonneutral as the PMD-Human severes (thick black line). SWISSPROTpolymorphisms (gray dotted line) are slightly more nonneutral than PMD-Human neutrals (solid gray line).
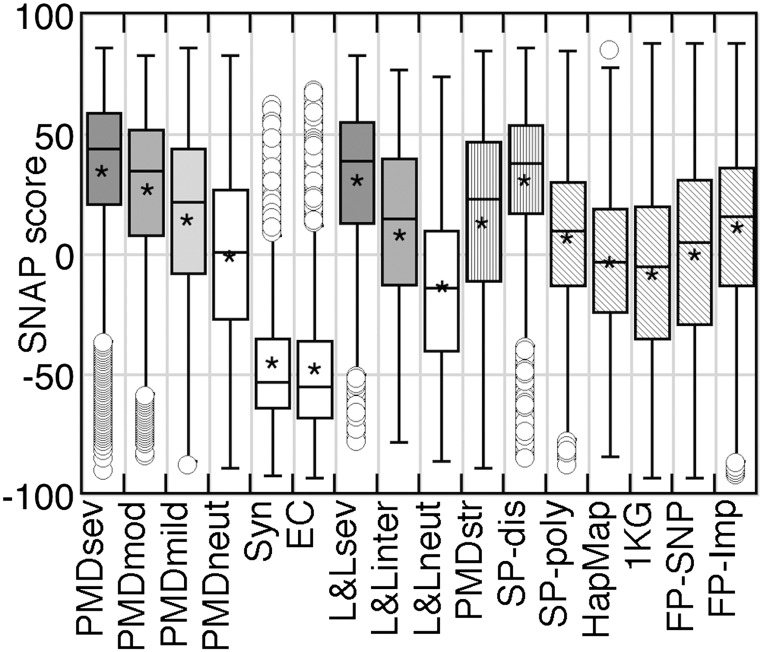
Fig. 2.
Summarizing the dataset distributions. The box-and-whisker plots and the distribution means capture the per-dataset trends. For each set, upper/lower box bounds represent the corresponding quartiles with the median shown as the crossbar. The “whiskers” of each box are the set’s minimum/maximum values with the outliers (points >1.5 times the interquartile distance away from the quartile bounds) not included in the calculations. A star in each box indicates the set mean. Solid gray and white boxes indicate functionally significant and neutral variants, respectively. Vertical shading indicates functionally unannotated variants, expected to be enriched for functional nonneutrals. Diagonal shading indicates that no inference of functional effect can be made for the set.
Prediction Scores Represent Functional Severity of Mutations.
Over 4,000 mutants in LacΙ repressor and over 2,000 in bacteriophage T4 lysozyme (“L&L”) were experimentally evaluated for functional impact (13, 14). The experimentally measured effect magnitudes agreed with the computational predictions (Figs. 1B and 2; “L&Lneutral” mean/median: −14/−14; “intermediate” mean/median: 11/15; “severe” mean/median: 30/39). Similarly for PMD, most variants experimentally shown to have an effect mapped into the range of predicted effects (scores, >0; Fig. 1C), with more severe mutations having higher (stronger) scores than less severe ones (Figs. 1C and 2; PMDsevere mean/median: 37/44; “PMDmoderate” mean/median: 28/35; and “PMDmild” mean/median: 16/22). Thus, although SNAP was not trained to differentiate levels of functional effect, the absolute value of the prediction score correlated with the experimentally observed severity of effect. If the experimental neutrals followed the same pattern, their score distribution would be significantly shifted to the left. The “PMDneutrals,” however, were split rather evenly into positive/negative predictions (Figs. 1C and 2; mean/median: −1/1). This result can be explained in two ways: (i) although SNAP succeeded for other neutral sets (EC, Synonymous, and L&Lneutral), it could have failed for PMDneutral; or (ii) the PMDneutral set might contain many incorrect annotations.
Other Methods Do Not Capture Effect Severity.
The two tools most commonly used to gauge functional effects of nsSNPs are SIFT (12) and PolyPhen-2 (9). Because PolyPhen-2 was developed for human proteins, we compared its performance to those of SNAP and SIFT using only the human protein variants in the PMD/EC set (“PMD/EC-Human”). Neither SIFT nor PolyPhen-2 was explicitly developed to reflect the effect severity. Still, the percentage of these methods’ nonneutral predictions (at default thresholds; SI Methods) varied with experimentally determined effect severity (e.g., PMD/EC-Human severes were predicted nonneutral more often than moderates and moderates more often than milds). The SIFT and PolyPhen-2 scores, however, were not informative of effect severity. In Fig. 1D, only the SNAP scores separate the gray (experimentally determined milds) and black (severes) distributions throughout the score range (i.e., SNAP scores correlate best with the observed effect severity).
Disease-Associated Mutations Have a Severe Effect on Function.
The prediction distribution for “SWISSPROTdisease” variants (Figs. 1E and 2; mean/median: 33/38) was very similar to that of PMD-Human severes [Kolmogorov–Smirnov distance (KSD) = 0.04; Fig. 1E and Table S2]. SNAP had not been trained on Swiss-Prot variants, i.e., these data illustrate the method performance for cases not seen before. In contrast, “SWISSPROTpolymorphism” predictions (Figs. 1E and 2; mean/median: 8/10) overlapped those for PMD-Human neutrals (Fig. 1E) and milds but distributed differently from either of these (KSD = 0.11 and 0.13, respectively). The SWISSPROTpolymorphisms have no established disease association. Thus, it is expected that these contain many functionally neutral and weakly nonneutral (nondisease) variants in agreement with our predictions (Fig. 1C).
Discussion
PMDneutrals Are Enriched in Variants with Weak Functional Effects.
Experimental annotation accuracy is especially important for large datasets, because misinterpreted results can lead to a waste of follow-up time and resources (15). Independent studies assessing annotations add important value. For example, we find that nearly half [44% (1139 of 2615); Dataset S1] of the variants annotated in the PMD as functionally neutral and evaluated for structural changes have been shown by experiments cited in the PMD to affect protein structure/stability. Because structural disturbance often changes function (16), these, and likely other, annotations of functional neutrality require deeper exploration. Moreover, the shape and position of the PMDneutral distribution suggests a set of weakly disruptive variants, closer to the PMDmilds (Fig. 1C) than to the true neutrals (“EC/Synonymous”; Fig. 1A and Tables S1 and S2).
The difference between the prediction distributions for the neutrals of the LacΙ repressor and bacteriophage T4 lysozyme set (L&Lneutrals) and that of the PMD is particularly interesting (Tables S1 and S2). The landscape of the L&L variants is complete in that all residues have been probed in the same manner. These homogeneous experimental annotations correspond to the ideal neutrals (EC/Synonymous) somewhat better (Fig. 2 and Tables S1 and S2). Additionally, the L&Lneutral and intermediate distributions are more distinct (KSD = 0.31) than the PMDneutrals and milds (KSD = 0.20; Fig. 1 B and C). Numerous different laboratories worked on PMDneutrals using different protocols, introducing more noise in annotations. Predictions for these show higher/more-nonneutral scores (Figs. 1C and 2). Selective mutagenesis and subjective annotations of experimentally observed function loss thus can miss real effects.
Mutation selection.
PMD entries summarize literature reports of mutations with experimentally measured functional significance. Mutations chosen for experimental evaluation, however, are rarely randomly selected. Rather, experimentalists generally suspect that these are disease related [e.g., genome-wide association studies (GWAS)], affect protein function (e.g., alanine scans for catalytic sites), or have structural significance (e.g., targeted mutagenesis in crystallography). Such hypothesis-driven results are enriched in effect variants compared with neutrals (PMD nonneutral:neutral ratio, ∼3:1; Table 1). In comprehensive mutagenesis studies, on the other hand, there is an over fourfold increase in the proportion of experimentally neutral mutations (L&L nonneutral:neutral ratio, ∼0.7:1). Removing this selection bias shifts the distribution for the L&Lneutrals to the left (more neutral) compared with PMDneutrals (Fig. 1 B and C).
Choice of functional properties for experimental follow-up.
The depth and comparability of experimental annotations is problematic. First, proteins may have many functional sites, and a given mutation may affect only some of these. Second, most experiments test only one particular reaction/functional assay. A variant neutral to one may affect another. Third, the threshold of identifying a variant as affecting function is not universal. A twofold decrease in e.g., binding may be deemed critical in one protein and accepted as within the wild-type range for another. Given these issues, we expect many PMDneutral variants will affect function.
Variants Affecting Structure Are Often Well Tolerated.
Many disease-causing mutations affect protein structure or stability (17). We found that of the disease-associated variants that were experimentally tested for structural impact, 75% (367 of 488 variants) have such an effect (Dataset S1). However, excluding variants that alter function in addition to affecting structure drastically decreases this percentage. Of 348 disease-associated variants experimentally evaluated for effects on both function and structure only, 20 (6%) are strictly structural, with no effect on function. One example is a histidine-to-tyrosine mutation at residue 107 of carbonic anhydrase 2, which is associated with mental retardation (18). The mutation renders the protein too unstable to purify (19), but the residual function of the mutant is reported to be unchanged: “kcat/Km [and the] pKa for the hydration of CO2 are identical to wild-type” (PMD entry A931076). More commonly, the variant affects both function and structure (64%) or only function with no effect on structure (27%). The finding that 11 (3%) of the disease-associated variants are experimentally annotated to have no effect on structure or function may point to a limited resolution of the experiments. Our numbers suggest that structurally disruptive variants often cause disease via changes to function. In other words, variants that disturb structure but leave function intact infrequently cause disease.
We have shown previously (20) that protein structure/stability change does not strictly correlate with function. Experimental evidence confirms this (Dataset S1) [i.e., of the PMD mutants with altered structure/stability and available function annotations, 22% (1139 of 5226) are experimentally functionally neutral and 41% have only mild effects (2158 of 5226)]. The predictions of structurally disruptive variants in PMD (“PMDstr”) distribute like those of weak effect variants [PMDmild; KSD = 0.03; Welch’s t test P value (WT-P) = 0.085]. Disease-associated nsSNPs, on the other hand, are enriched in variants predicted to severely disrupt function (21). SWISSPROTdisease mutations (Fig. 1E) distribute similarly to PMD-Human severes (KSD = 0.04). The difference between the SWISSPROTdisease and PMDstr functional effect distributions (KSD = 0.21; WT-P = 7.35 × 10−306) confirms the idea that structurally significant mutations, unless specifically disruptive of function, rarely cause disease.
Many Variants in Genomes of Healthy Individuals Predicted to Impact Function.
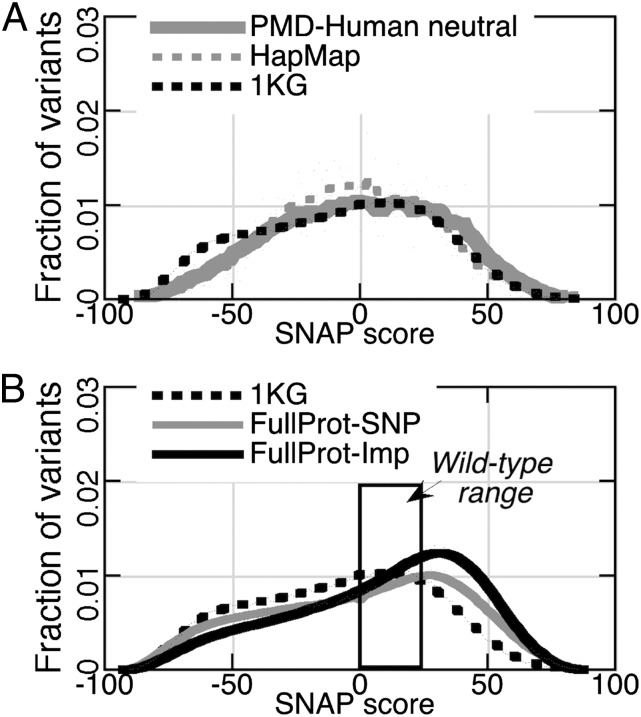
SWISSPROTpolymorphisms (Fig. 1E) are slightly more functionally disruptive than PMD-Human neutrals (KSD = 0.11). As polymorphism annotations are subjective, this needs additional confirmation. We predicted the functional effects for variants in apparently healthy individuals in both the 1000 Genomes (“1KG”) (22) data and the subset of HapMap (Haplotype Map) (23) variants (“HapMap”) that exist in all probed populations (SI Methods) (24). As expected from their healthy human origin, both sets contain few predicted severely nonneutral mutations. However, other studies (10) and experimental findings (PMD) identify at least some nonneutrals in the 1KG data. HapMap’s distribution (Figs. 3A and 2; mean/median: −3/−3) is similar to that of the PMD-Human neutrals (KSD = 0.07). That is, we find that common SNPs (>10% frequency) are only a little more likely (54%) to be functionally identical to the reference than different from it (46%). The nonneutral common SNPs are either weakly advantageous or weakly deleterious and subject to balancing selection. The neutrals are maintained in a population as long as no negative selection pressure is applied. These findings are consistent with recent work that highlights the modest contribution of common variants to complex traits (25). Compared with HapMap, the 1KG distribution is very different in shape and shifted more to the left (neutral; KSD = 0.89; mean/median: −8/−5; Figs. 3A and 2). This difference is not surprising because HapMap is a unique and small subset of all human variants. As a nonneutral percentage of the total, however, the two sets are similar. The rare variants of healthy individuals (bulk of 1KG) are also only slightly more likely to be functionally identical to reference (at SNAP score, 0; 55%) than different from it (45%). This is also expected, because severely deleterious variants would not be present in a healthy population, and advantageous variants would spread to become common.
Fig. 3.
Distribution of human variant functional effects. A and B were created as in Fig. 1. (A) Both 1KG (black dots) and HapMap (gray dots) distributions are similar to PMD-Human neutrals (gray line). (B) FullProt-Imp (SNP impossible) mutations (black line) are, on average, more deleterious than FullProt-SNP (SNP possible) ones (gray line). 1KG variants (black dots) do not encompass the whole diversity of SNP possible mutants. This suggests a physiologically acceptable range of nonidentical protein functionality (the neutral range, black box): 0 < SNAP ≤ 23, where mildly nonneutral SNPs are still retained by the human proteome.
“Wild type” as applied to individual proteins refers to the biochemical properties of a reference sequence, whereas “wild type” at the level of the whole organism’s physiology refers to a range of protein functionality consistent with the absence of disease. We show that proteins translated from specific allele sequences in the genomes of healthy individuals are often functionally different from reference sequences. Currently, the database reference protein activity is defined as wild type. If an individual’s specific protein has a different activity, it is not wild type. Our results thus describe the existence of a physiologically wild-type range, a selectively neutral range of non–wild-type protein activity that presents as a wild-type phenotype on the whole-organism level.
Genes Under Selective Pressure Diversify but Are Predicted to Maintain Functionality.
The evolution of the genetic code is constrained by a balance between robustness and changeability (26). That is, protein-coding sequences balance the ability to tolerate and/or potentially put to use genetic “spelling errors.” It is accepted that the nucleotide-sequence similarity of the codons encoding biochemically similar amino acids has evolved to allow for phenotypic flexibility (27) (i.e., nucleotide variations have minimal effect on the gene product). Our results confirm this view. The set of all single amino acid substitutions possible via a SNP (“FullProt-SNP”) is overall predicted as more functionally neutral than the SNP-impossible set (“FullProt-Imp”) in the same proteins (Figs. 2 and 3B). Additionally, we show that the human proteome is actually intolerant of a significant portion of even the SNP-possible variants. The distribution of the real nsSNPs in healthy human genomes (1KG; Fig. 3B) is more neutral than either of the FullProt sets (i.e., we do not observe a large fraction of the possible, albeit functionally nonneutral, nsSNPs). This finding is in line with the notion of variant purifying selection (28, 29).
A similar observation can be made using PolyPhen-2. “PolyPhen-natural” variants (all known human nsSNPs, SI Methods) exhibit a roughly 5:3 ratio of neutral to nonneutral predictions, even though some of these may be disease associated. On the other hand, “PolyPhen-SNP” mutants (all SNP-possible mutations in all human genes) display a 5:9 ratio instead: a threefold increase in nonneutral SNPs compared with the naturally occurring set.
SNPs affecting functionally critical sites, such as catalytic residues or binding hotspots, are rarely tolerated. Functionally essential (and ultraconserved) proteins (e.g., ubiquitin or histones) contribute to the lack of allowed variability. However, this relatively small set of residues should be easily counter balanced in length by the regions of low conservation present in almost every protein. The neutral lean of the 1KG scores compared with the FullProt-SNP distribution suggests that not all functional variation is acceptable on the organism level. The overlap between the two distributions in the region of functional significance (SNAP score, >0) constitutes the physiologically wild-type range of protein activity, the territory in the protein functional landscape where activity is not equivalent to reference but not yet different enough to cause whole-organism level changes. To estimate where this transition occurs, we observe that more SNPs with scores >23 are excluded from the proteomes by selective pressure than are acceptable (intersection of FullProt-SNP with 1KG; Fig. 3B). Over 70% (3705 of 5194) of PMD disease mutants score ≥23. On the other hand, ∼79% of all human variants (1KG) are predicted below this cutoff and, as such, are in the physiologically wild-type range of protein function. The variants predicted to affect function in any way (0 < score ≤ 23; ∼23% of 1KG) are still not entirely “safe” because their combinations may contribute to complex disease. In a sense, the concept of wild type is, thus, redefined from the exact level of functionality of “the most common allele in a population” to a wild-type range of activity that is tolerated without significant phenotypic changes on the organismal level.
Limitations and Potential Pitfalls of Large-Scale Computational Variant Studies.
Although, in terms of time and effort required for large-scale analyses, computational predictions are preferential to detailed experiments, the latter are significantly more accurate. SNAP is ∼80% accurate in identifying binary (neutral/nonneutral) variant functional effects. This incomplete resolution could affect our conclusions. Here, we consider the potential problems of our analysis.
First, we note that in prior work (8), we showed that SNAP is as accurate for variants it has never seen as for those that it has seen in training. Here, we demonstrate SNAP’s ability to identify new neutral variants: the distribution of Synonymous predictions is very similar to our best benchmark performance on the EC set (KSD = 0.08). Second, we compare score distributions over large variant sets. Arguably, because in a comparison, both distributions are subject to the same errors, their difference more accurately represents reality than either of the distributions alone (i.e., even if SNAP was 95% accurate per prediction, the difference between any two prediction distributions would likely remain the same). Note that comparison-based inferences are subject to the statistical testing limitations. For this reason, we used different statistical methods to examine distribution differences (Tables S1 and S2). Third, computational evidence suggests that experimental annotation of neutrality is imprecise [i.e., the distribution of PMDneutrals is different from that of other neutrals (Tables S1 and S2) but similar to that of FullProt-SNP (KSD = 0.06; WT-P = 0.44)]. Biologically, this concordance of the distributions of neutral variants with that of all SNP-possible variants is unlikely. Furthermore, at the SNAP-defined physiological wild-type cutoff of 23, over 72% of the PMDneutrals are correctly identified. This suggests that experimental evaluations of variant neutrality often report effects on the overall organism phenotype instead of on the specific protein functions. Finally, although it is theoretically possible that SNAP misannotates PMDneutrals more than other sets, there is no way of confirming or refuting this without extensive experimental follow up of thousands of variants.
Functional Neutrality Is Nondiscreet.
Here, three conclusions are salient:
1) At least a fifth (21–45%, at the physiological wild-type cutoff of 23 and the default neutral cutoff of 0, respectively) of the variants in the genomes of healthy individuals render affected proteins functionally different from reference genome proteins (i.e., individuals differ from each other via an extensive collection of small changes). Assessing the “combinatorial load” of these nonneutral variants in relevant pathways will likely contribute to understanding the genetics of complex disease.
2) Common variants in healthy individuals, on average, affect protein function as much as the rare SNPs. This is not surprising: to spread through the population common SNPs should be either advantageous with respect to reference or selectively neutral. An advantageous variant, by definition, significantly affects function. Selectively neutral variants can be within the physiologically wild-type range if they carry no (dis)advantage to the organism as a whole, even if they are not neutral with respect to the individual protein's reference function. Nonneutral rare variants, however, which have an effect well beyond the physiologically wild-type range, are arguably infrequent in healthy genomes. Rare variants are enriched in functional nonneutrals compared with a neutral model of evolution (25). We suggest that these are often of weak effect, may have entered the population recently (30), and are, thus, “fuel for evolution.” The fraction of common variant nonneutrals in healthy individuals (46% nonneutrals in HapMap) is at least as high as of rare variants (45% nonneutrals in 1KG) because (i) selection pressure causes advantageous variants to be fixed (become common) quickly, (ii) neutrals and weak nonneutrals are normally fixed at about the same rate via genetic drift (31), and (iii) in the presence of an evolutionary bottleneck, weakly nonneutral variants contribute to species fitness (chance of survival) (30). In individuals affected by complex diseases, which are often attributable to additive effects of many variants, the fraction of nonneutral rare variants is likely higher than that of nonneutral common variants (30).
3) Regardless of SNAP’s accuracy in neutral/nonneutral classification of mutations, the similarity of the 1KG, HapMap, and PMDneutral distributions suggests that most nsSNPs in the genome of a healthy individual would be deemed experimentally neutral given current experimental methods. However, it is likely that many of these mutations are actually weakly nonneutral with respect to reference protein function. Although indicative of a certain deficiency in experimental resolution, this finding is consistent with the level of observed whole-organism tolerance to protein functional changes within the physiologically wild-type range.
Conclusions
We show that healthy individuals differ by a large collection of variants that are often invisible to current state-of-the-art “wet lab” experimentation. The variants are within the organism-level physiologically wild-type range even as they change specific protein function. In nature, they are fuel for evolution. For humans, they may be a key to development of personalized medicine. Complex disease analysis of the future needs to account for both the lower-than-expected sensitivity of the experimental methods to the effects of polymorphisms and for the inherent variation of functionality in the individual genomes.
Methods
Dataset extractions (Table 1, Table S3, and Dataset S1), comparison methods (Tables S1 and S2), and prediction runs are described in SI Methods. Note that the KSD is computed on discreet distributions in the presence of ties and is, thus, approximate. It is used here solely for comparison of distribution-pair distances and does not indicate the statistical significance of distribution similarities (SI Methods).
Supplementary Material
Acknowledgments
We thank G. Yachdav, C. Schaefer, and L. Kajan [all from Technical University of Munich (TUM)] for computational help. We are grateful to N. Samusik (Stanford), T. Goldberg (TUM), and C. Zhu, B. Jelen, and S. Senn (all from Rutgers) for many important discussions. We also thank all of those who deposited their experimental results into public databases. The work of Y.B. is supported by School of Environmental and Biological Sciences (SEBS), Rutgers University start-up funds. B.R. is supported by the Alexander von Humboldt Foundation. P.C.K. acknowledges support by SEBS and an anonymous private source.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1216613110/-/DCSupplemental.
References
- 1.Winkelmann BR, Hager J. Genetic variation in coronary heart disease and myocardial infarction: Methodological overview and clinical evidence. Pharmacogenomics. 2000;1(1):73–94. doi: 10.1517/14622416.1.1.73. [DOI] [PubMed] [Google Scholar]
- 2.Pauling L, et al. Sickle cell anemia, a molecular disease. Science. 1949;109(2835):443. [PubMed] [Google Scholar]
- 3.Smyth DJ, et al. A genome-wide association study of nonsynonymous SNPs identifies a type 1 diabetes locus in the interferon-induced helicase (IFIH1) region. Nat Genet. 2006;38(6):617–619. doi: 10.1038/ng1800. [DOI] [PubMed] [Google Scholar]
- 4.Kawabata T, Ota M, Nishikawa K. The Protein Mutant Database. Nucleic Acids Res. 1999;27(1):355–357. doi: 10.1093/nar/27.1.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.UniProt Consortium Reorganizing the protein space at the Universal Protein Resource (UniProt) Nucleic Acids Res. 2012;40(Database issue):D71–D75. doi: 10.1093/nar/gkr981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Riazanov A, Laurila JB, Baker CJ. Deploying mutation impact text-mining software with the SADI Semantic Web Services framework. BMC Bioinformatics. 2011;12(Suppl 4):S6. doi: 10.1186/1471-2105-12-S4-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goldberg T, Hamp T, Rost B. LocTree2 predicts localization for all domains of life. Bioinformatics. 2012;28(18):i458–i465. doi: 10.1093/bioinformatics/bts390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bromberg Y, Rost B. SNAP: Predict effect of non-synonymous polymorphisms on function. Nucleic Acids Res. 2007;35(11):3823–3835. doi: 10.1093/nar/gkm238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Adzhubei IA, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7(4):248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xue Y, et al. 1000 Genomes Project Consortium Deleterious- and disease-allele prevalence in healthy individuals: Insights from current predictions, mutation databases, and population-scale resequencing. Am J Hum Genet. 2012;91(6):1022–1032. doi: 10.1016/j.ajhg.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kondrashov AS. Contamination of the genome by very slightly deleterious mutations: Why have we not died 100 times over? J Theor Biol. 1995;175(4):583–594. doi: 10.1006/jtbi.1995.0167. [DOI] [PubMed] [Google Scholar]
- 12.Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31(13):3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Markiewicz P, Kleina LG, Cruz C, Ehret S, Miller JH. Genetic studies of the lac repressor. XIV. Analysis of 4000 altered Escherichia coli lac repressors reveals essential and non-essential residues, as well as “spacers” which do not require a specific sequence. J Mol Biol. 1994;240(5):421–433. doi: 10.1006/jmbi.1994.1458. [DOI] [PubMed] [Google Scholar]
- 14.Rennell D, Bouvier SE, Hardy LW, Poteete AR. Systematic mutation of bacteriophage T4 lysozyme. J Mol Biol. 1991;222(1):67–88. doi: 10.1016/0022-2836(91)90738-r. [DOI] [PubMed] [Google Scholar]
- 15. (2012) Must try harder. Nature 483(7391):509. [DOI] [PubMed]
- 16.Sánchez IE, Tejero J, Gómez-Moreno C, Medina M, Serrano L. Point mutations in protein globular domains: Contributions from function, stability and misfolding. J Mol Biol. 2006;363(2):422–432. doi: 10.1016/j.jmb.2006.08.020. [DOI] [PubMed] [Google Scholar]
- 17.Wang Z, Moult J. SNPs, protein structure, and disease. Hum Mutat. 2001;17(4):263–270. doi: 10.1002/humu.22. [DOI] [PubMed] [Google Scholar]
- 18.Soda H, et al. A point mutation in exon 3 (His 107—>Tyr) in two unrelated Japanese patients with carbonic anhydrase II deficiency with central nervous system involvement. Hum Genet. 1996;97(4):435–437. doi: 10.1007/BF02267062. [DOI] [PubMed] [Google Scholar]
- 19.Tu C, Couton JM, Van Heeke G, Richards NG, Silverman DN. Kinetic analysis of a mutant (His107—>Tyr) responsible for human carbonic anhydrase II deficiency syndrome. J Biol Chem. 1993;268(7):4775–4779. [PubMed] [Google Scholar]
- 20.Bromberg Y, Rost B. Correlating protein function and stability through the analysis of single amino acid substitutions. BMC Bioinformatics. 2009;10(Suppl 8):S8. doi: 10.1186/1471-2105-10-S8-S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schaefer C, Bromberg Y, Achten D, Rost B. Disease-related mutations predicted to impact protein function. BMC Genomics. 2012;13(Suppl 4):S11. doi: 10.1186/1471-2164-13-S4-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abecasis GR, et al. 1000 Genomes Project Consortium A map of human genome variation from population-scale sequencing. Nature. 2010;467(7319):1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.International HapMap Consortium The International HapMap Project. Nature. 2003;426(6968):789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 24.Schaefer C, Meier A, Rost B, Bromberg Y. SNPdbe: Constructing an nsSNP functional impacts database. Bioinformatics. 2012;28(4):601–602. doi: 10.1093/bioinformatics/btr705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kiezun A, et al. Exome sequencing and the genetic basis of complex traits. Nat Genet. 2012;44(6):623–630. doi: 10.1038/ng.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maeshiro T, Kimura M. The role of robustness and changeability on the origin and evolution of genetic codes. Proc Natl Acad Sci USA. 1998;95(9):5088–5093. doi: 10.1073/pnas.95.9.5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Freeland SJ, Wu T, Keulmann N. The case for an error minimizing standard genetic code. Orig Life Evol Biosph. 2003;33(4-5):457–477. doi: 10.1023/a:1025771327614. [DOI] [PubMed] [Google Scholar]
- 28.Fay JC, Wyckoff GJ, Wu CI. Positive and negative selection on the human genome. Genetics. 2001;158(3):1227–1234. doi: 10.1093/genetics/158.3.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bustamante CD, et al. Natural selection on protein-coding genes in the human genome. Nature. 2005;437(7062):1153–1157. doi: 10.1038/nature04240. [DOI] [PubMed] [Google Scholar]
- 30.Fu W, et al. NHLBI Exome Sequencing Project Analysis of 6,515 exomes reveals the recent origin of most human protein-coding variants. Nature. 2013;493(7431):216–220. doi: 10.1038/nature11690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kimura M, Ohta T. The average number of generations until fixation of a mutant gene in a finite population. Genetics. 1969;61(3):763–771. doi: 10.1093/genetics/61.3.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stineman RW. A consistently well behaved method of interpolation. Creative Computing. 1980;6(7):54–57. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.