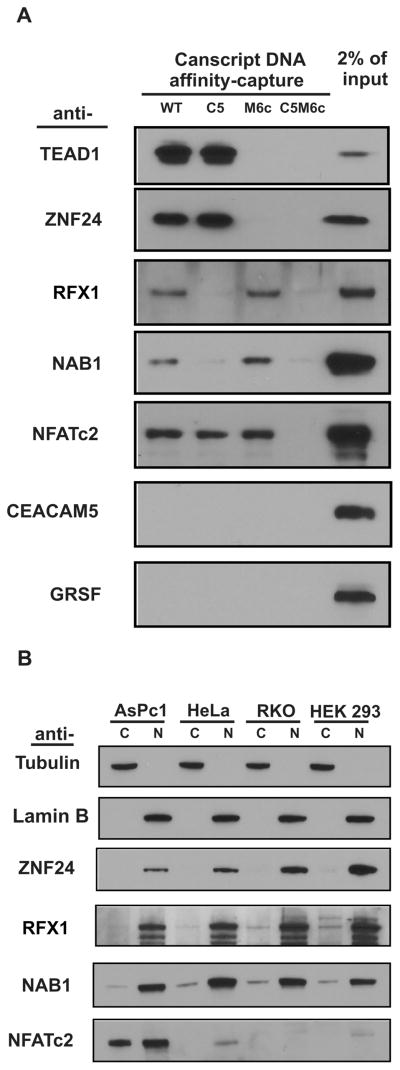
Figure 4.
Verification of hits identified by SD-MS. (A) Seven of nine hits listed in Table 1 were examined by immunoblot following DNA affnity-capture. Single mutant (C5 and M6c) CanScript ×3 probes were used in parallel to attribute which binding motif was responsible for selected hits. The protocol was as described in Figure 2. (B) Expression pattern and subcellular distribution of ZNF24, RFX1, NAB1 and NFATc2 in MSLN-positive (AsPc1 and HeLa) and -negative (RKO and HEK293) cell lines. Alpha-tubulin was used as a cytoplasmic marker (C) and lamin B as a nuclear marker (N).