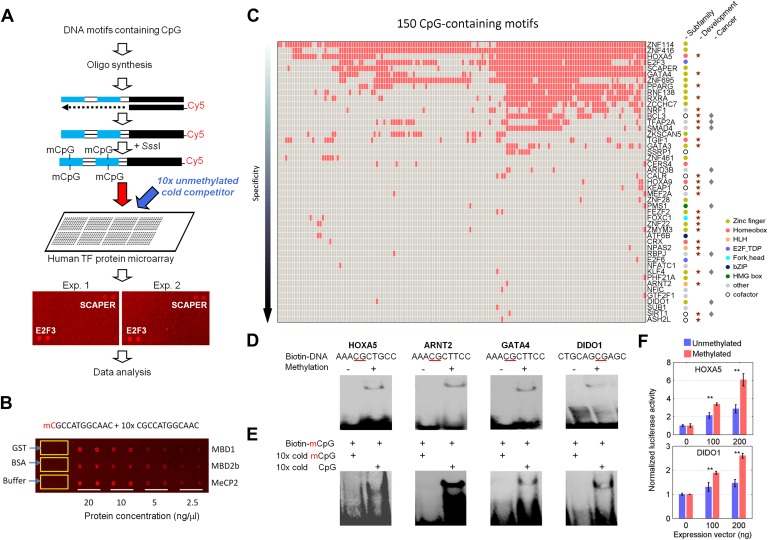
Figure 1. Protein microarray-based approach identified mCpG-dependent DNA-binding activity among human TFs and cofactors.
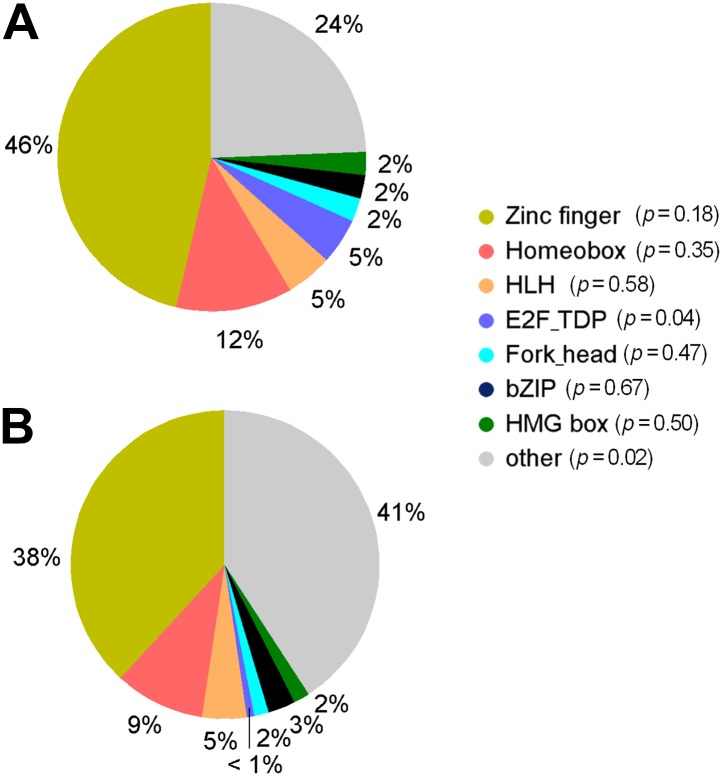
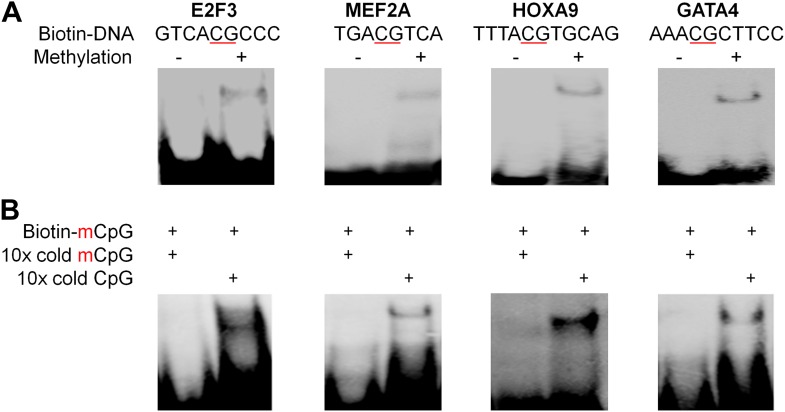
(A) A competition assay was used to identify proteins that preferentially bind to methylated DNA motifs. SCAPER (S-phase cyclin A-associated protein in the ER) and E2F3 (E2F transcription factor 3) were shown here as two examples of methylated DNA-binding proteins. (B) A proof-of-principle assay was conducted using known methylated DNA-binding proteins on a pilot protein microarray. (C) Binding profiles of 41 TFs and 6 co-factors against 150 of the 154 tested methylated DNA motifs are summarized in the interaction map. TFs are color-coded based on the subfamilies. (D) EMSA assays validated DNA-binding activity for four selected TF candidates. Representative images from three independent experiments with similar results are shown. (E) Competition EMSA assays confirmed mCpG-dependent DNA-binding activities. As expected, 10-fold unlabeled, methylated DNA motif readily abolished the protein–DNA complex formation of the tested TFs with the biotinylated and methylated DNA motifs (Lane 1 in each image). However, 10-fold cold unmethylated DNA counterparts could not compete off methylated DNA binding, consistent with the protein microarray results. (F) HOXA5 and DIDO1 showed mCpG-dependent activation of luciferase activity in GT1-7 cells. Values represent mean ± SD (n = 3; **: p<0.01; t-test).