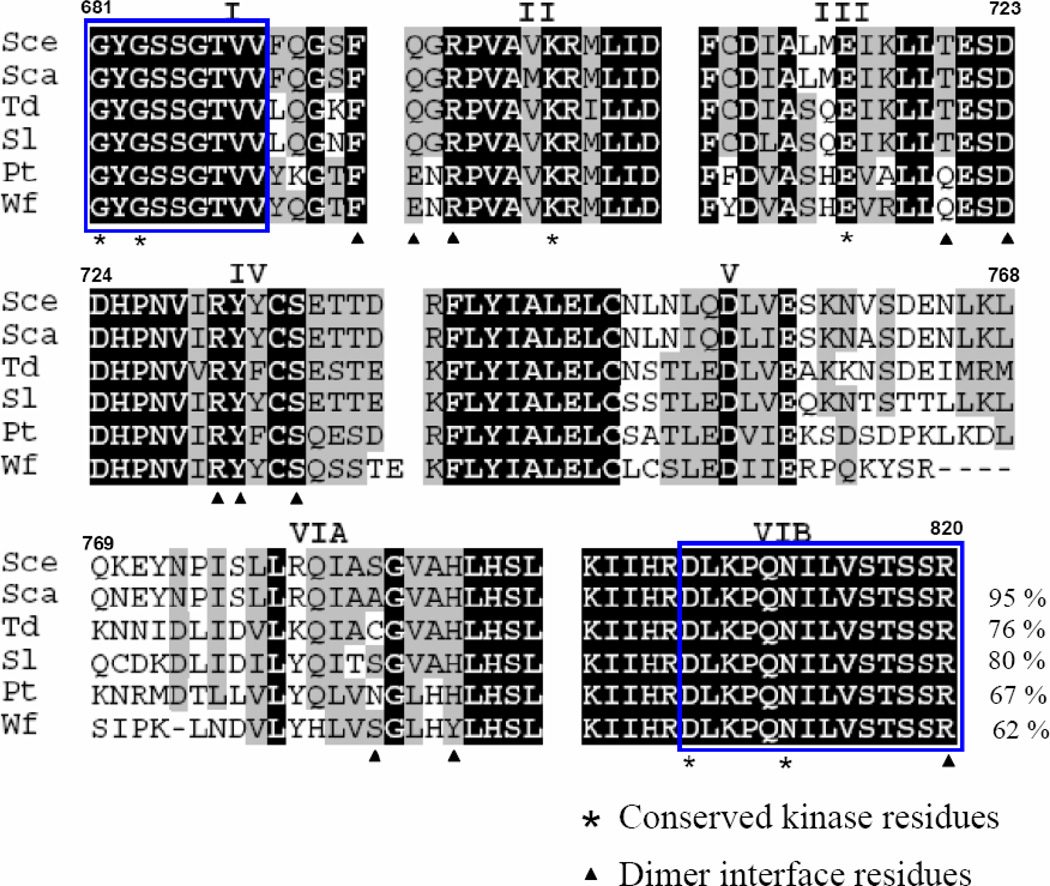
Fig 2. Amino acid alignment of Ire1 homologues.
Partial deduced amino acid sequences of Ire1 homologues from Saccharomyces carlsbergensis (accession number FN677757), Saccharomycodes ludwigii (accession number FN677758), Pachysolen tannophilus (accession number FN677759), Torulasopora delbrueckii Wf (accession number FN677760) and Wickerhamia fluorescens (accession number FN677761) were aligned to Saccharomyces cerevisiae Ire1. Roman numerals indicate kinase subdomains. Blocks represent the position of degenerate primers. Dark shading indicates identical residues. Light shading indicates conserved residues. Numbers indicate amino acid residues according to S. cerevisiae Ire1(Sce). Percent sequence identities to S. cerevisiae Ire1 are shown in the last line.