Figure 3.
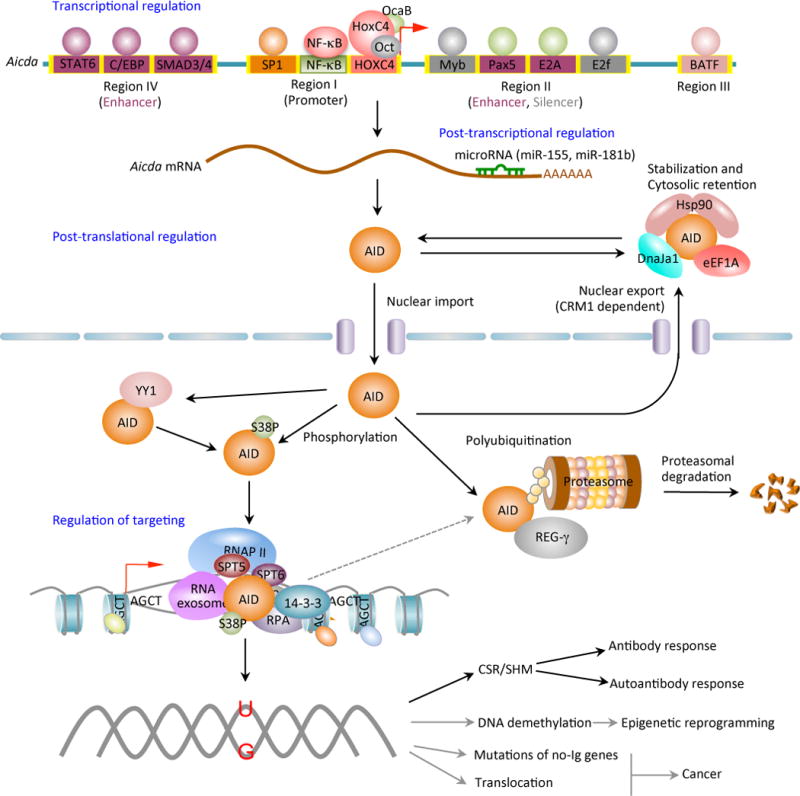
AID expression and activity are tightly regulated at the levels of transcription, post-transcription, post-translation (including nuclear/cytoplasmic distribution and stability) and enzymatic function. Four distinct DNA regions (regions I to IV) of the AID gene (Aicda) locus containing binding sites for multiple transcription factors have been shown to regulate Aicda expression. Region I function as promoter containing the binding sites for HoxC4/Oct and NF-κB/Sp1/Sp3, which can be induced by activate the Aicda promoter. In resting naive and memory B cells, as well as in non-B cells, silencer elements in region II bind the repressor proteins E2f and c-Myb to counter the activity of the transcriptional activators. Stimulation of B cells with the primary inducing stimuli and cytokines that promote CSR induce activation signals through region IV enhancer in collaboration with the intronic enhancer in region II can overcome the effect of the region II silencer. After transcription, the Aicda mRNA can be negatively regulated by microRNAs, including miR-155 and miR-181b, which specifically bind to the conserved target sites on the 3’ UTR of Aicda mRNA. The AID protein undergoes a series of post-translational modifications, such as dimerization/oligomerization, nuclear/cytoplasmic translocation, phosphorylation and polyubiquitination, all of which are important for AID activity. Most nuclear AID is either degraded or exported back to the cytoplasm. Only a small proportion of AID molecules are targeted onto DNA at Ig or non-Ig loci by its co-factors. Nuclear AID is constantly targeted to (protein degrading) proteasomes by ubiquitin-dependent and -independent pathways. The ubiquitin-independent pathway relies on the nuclear protein REG-γ, a proteasomal activator that is ubiquitin- and ATP-independent. AID preferentially deaminates single-strand DNA, which emerges from transcription by RNA Pol II and depends on histone modifications in the transcribed locus. AID can be recruited to the open DNA before or during transcription. In CSR, AID recruitment to S regions occurs with interaction of RNA Pol II and AID-interacting factors, such as Spt5, Spt6, PTBP2, RNA exome and 14-3-3 adaptor proteins, which would form a macromolecular complex. AID is enriched and stabilized on the targeted DNA by 14-3-3 adaptor proteins, which access the same S regions as the transcription machinery owing to their open chromatin state. These 14-3-3 adaptors are recruited and/or stabilized through interactions with 5′-AGCT-3′ repeats and possibly by H3K9acS10ph. The RNA exosome also interacts with AID and allows AID to deaminate both the transcribed and the non-transcribed DNA strand in the S regions undergoing transcription. AID deaminate dCs into dUs to yield dU:dG mismatches. Resolution of these lesions can lead to different physiologic or pathologic outcomes.
