Transcriptomic analyses show that apomixis-initiating cells embark on an embryo sac program prior to nuclear division.
Abstract
Hieracium praealtum forms seeds asexually by apomixis. During ovule development, sexual reproduction initiates with megaspore mother cell entry into meiosis and formation of a tetrad of haploid megaspores. The sexual pathway ceases when a diploid aposporous initial (AI) cell differentiates, enlarges, and undergoes mitosis, forming an aposporous embryo sac that displaces sexual structures. Embryo and endosperm development in aposporous embryo sacs is fertilization independent. Transcriptional data relating to apomixis initiation in Hieracium spp. ovules is scarce and the functional identity of the AI cell relative to other ovule cell types is unclear. Enlarging AI cells with undivided nuclei, early aposporous embryo sacs containing two to four nuclei, and random groups of sporophytic ovule cells not undergoing these events were collected by laser capture microdissection. Isolated amplified messenger RNA samples were sequenced using the 454 pyrosequencing platform and comparatively analyzed to establish indicative roles of the captured cell types. Transcriptome and protein motif analyses showed that approximately one-half of the assembled contigs identified homologous sequences in Arabidopsis (Arabidopsis thaliana), of which the vast majority were expressed during early Arabidopsis ovule development. The sporophytic ovule cells were enriched in signaling functions. Gene expression indicative of meiosis was notably absent in enlarging AI cells, consistent with subsequent aposporous embryo sac formation without meiosis. The AI cell transcriptome was most similar to the early aposporous embryo sac transcriptome when comparing known functional annotations and both shared expressed genes involved in gametophyte development, suggesting that the enlarging AI cell is already transitioning to an embryo sac program prior to mitotic division.
Some Hieracium subgenus Pilosella species form seed via sexual reproduction. Others are facultative for apomixis, where the majority of seed is formed via an asexual pathway and therefore genetically identical, while a small proportion of seed is derived via sexual reproduction. Female gametophyte (or embryo sac) development in ovules of sexual Hieracium species occurs via the most common pathway observed in angiosperms (Drews and Koltunow, 2011). It initiates with megasporogenesis, a process requiring diploid megaspore mother cell (MMC) differentiation and subsequent MMC meiosis to produce a tetrad of four haploid megaspores. Three of these megaspores undergo cell death. The surviving or functional megaspore (FM) undergoes megagametogenesis, characterized by three rounds of syncytial nuclear mitosis, followed by cellularization and differentiation to produce the mature female gametophyte. Six cells in the female gametophyte contain a haploid nucleus including the egg cell, two synergids, and three antipodal cells, while the central cell contains two haploid nuclei that fuse prior to double fertilization. Fertilization of the haploid egg and the diploid central cell in the female gametophyte by haploid male sperm cells triggers formation of the embryo and endosperm compartments of the seed respectively (Fig. 1A, yellow; Koltunow et al., 1998).
Figure 1.
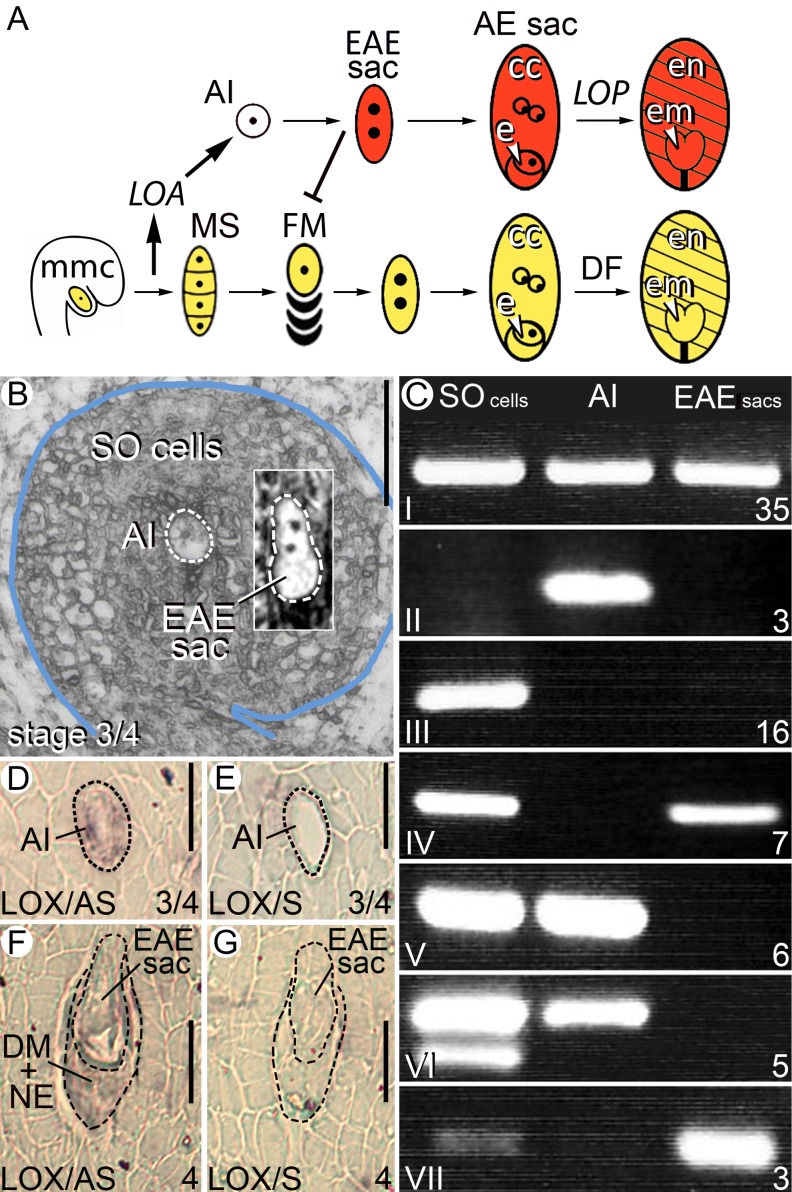
Sexual and apomictic pathways in H. praealtum, laser-captured cell types and analysis of gene expression. A, In apomictic H. praealtum, MMC meiosis and tetrad formation during sexual reproduction (yellow) activates the LOA locus leading to AI cell differentiation and EAE sac formation (red). Sexual development ceases in approximately 97% of ovules as the EAE sac displaces the sexual pathway generally at FM selection. A mature aposporous embryo sac develops with a diploid egg (e) and two diploid central cell (cc) nuclei. The LOP locus triggers fertilization-independent embryo (em) and endosperm (en) formation. Incomplete penetrance of LOA and LOP in approximately 3% of ovules results in haploid embryo sac formation, and double fertilization (DF) is required for seed initiation in gametophytes where LOP is lost by segregation. B, The three cell types isolated by LCM. Uninucleate AI cells and EAE sacs containing two to four nuclei were dissected in addition to SO cells collected from random locations in the ovule section. Bar = 50 µm. C, Seven classes of gene expression observed in the aRNA from the three laser-captured samples using RT-PCR to detect a suite of low-level-expressed ovary test genes. The number of genes in each class is indicated on the bottom right. D to G, In situ analysis in H. praealtum ovules of a LOX gene, one of three AI cell-expressed genes from class II in C. Antisense (AS) probes were used in D and F and control sense (S) probes in E and G. In situ analysis of the other two genes in H. praealtum and in sexual H. pilosella are shown in Supplemental Figure S3. Bar = 20 µm.
In apomictic Hieracium subgenus Pilosella species, the MMC initiates and completes meiosis as observed in sexual species. The meiotic events of megasporogenesis are essential for apomixis initiation in Hieracium piloselloides and are thought to activate function of the dominant, hemizygous LOSS OF APOMEIOSIS (LOA) locus conserved in three apomictic subgenus Pilosella species. Information and/or structure at the LOA locus enables differentiation of aposporous initial (AI) cells, their mitotic development into aposporous embryo sacs, and subsequent suppression of the adjacent sexual pathway (Koltunow et al., 2011a; Okada et al., 2011). Multiple AI cells stochastically differentiate near cells undergoing megasporogenesis. Usually, one AI cell undergoes mitosis to form the early aposporous embryo (EAE; containing two to four nuclei) sac, which enlarges toward the megaspores and/or FM concurrent with displacement and termination of the sexual pathway (Fig. 1A, red; Koltunow et al., 2011a). A mature diploid aposporous embryo sac forms after additional mitoses, positionally substituting for the sexual embryo sac. Action of the independent LOSS OF PARTHENOGENESIS (LOP) locus in Hieracium praealtum induces fertilization-independent proliferation of the egg and central cell, giving rise to seeds containing embryos with a maternal genotype (Fig. 1A, red; Koltunow et al., 2011a). Deletion of the LOA and LOP loci by irradiation in H. praealtum results in loss of apomixis with coincident reversion of the plant to sexual reproduction. Apomixis in H. praealtum is therefore superimposed on sexual reproduction and does not result from a mutational inactivation of the sexual process (Catanach et al., 2006; Koltunow et al., 2011a). Incomplete functional penetrance of LOA and LOP loci is thought to give rise to the low levels of sexual progeny in these facultative apomicts (Koltunow et al., 2011a).
LOA and LOP loci are hypothesized to induce a modified sexual pathway in the AI cell and its descendant structures where both meiosis and fertilization are avoided. This notion is derived from earlier expression analyses using a small set of reproductive tissue marker genes in transgenic sexual and apomictic Hieracium. Sexual and apomictic reproductive pathways showed convergence and common expression regulation, from the first nuclear division of the AI cell in apomictic H. piloselloides and Hieracium aurantiacum, and the first nuclear division of the FM as it entered megagametogenesis in sexual H. pilosella. These analyses suggested that once the AI cell undergoes its first nuclear division, it is committed to a megagametogenic program (Tucker et al., 2003).
The LOA-induced gene expression programs contributing to AI cell specification, differentiation, and growth are unknown, as is the functional identity of the AI cell before it undergoes nuclear division. Hypothetically, the AI cell may differentiate as an ectopic MMC or have FM identity upon differentiation, as both are gametic precursor cells. Current evidence in apomictic Hieracium spp. concerning both possibilities is inconclusive. Cytological studies using aniline blue indicate the AI cell lacks detectable callose in its cell wall, similar to that of FMs. This contrasts with callose-rich cell walls of MMCs in examined sexual and apomictic Hieracium subgenus Pilosella species (Tucker et al., 2001). The Arabidopsis (Arabidopsis thaliana) SPOROCYTELESS:GUS marker construct is expressed in the MMC of sexual and apomictic Hieracium spp. but not in AI cells (Tucker et al., 2003). The DISRUPTION OF MEIOTIC CONTROL1 (DMC1) gene, which is required for interhomolog recombination during meiosis (Couteau et al., 1999), is expressed in the MMCs of sexual and apomictic Hieracium spp. but is undetectable in AI cells via in situ hybridization (Okada et al., 2007). While these data imply the AI cell is unlikely to have MMC identity, and more likely to differentiate with FM identity, the expression of other meiosis genes has not been examined. Thus, the possibility that the meiotic pathway may initiate and deviate at other stages in developing AI cells cannot currently be excluded. The possibility that in apomictic Hieracium spp. AI cells possess an FM-type identity is also unresolved due to the limited availability of megaspore-specific markers to test this. The Arabidopsis FM marker pFM1 (Acosta-García and Vielle-Calzada, 2004) has been introduced to sexual and apomictic Hieracium spp. but is not expressed in ovules (Koltunow et al., 2011a, 2011b). Collectively, these analyses highlight the current paucity of cell type-specific markers and ovule EST sequence information pertaining to early apomictic development in aposporous Hieracium spp.
In this study, we used laser capture microdissection (LCM) and 454 pyrosequencing of the isolated and amplified RNA (aRNA) to examine gene expression in enlarging AI cells and EAE sacs compared with surrounding ovule cells during apomictic initiation in H. praealtum. Indicative roles of each cell type were established through comparisons of expressed sequences across all three cell types and analyses of sequence annotations derived through homology to known genes and protein motifs. These analyses have revealed close functional similarity between AI cells and EAE sacs and significant enrichment of signaling functions in surrounding sporophytic ovule (SO) cells, which may impact on apomixis initiation and development in aposporous H. praealtum.
RESULTS
RNA Isolated from Laser-Captured Cell Types Maintains Representation of Low-Abundance Transcripts
LCM was used to isolate specific ovule cell types to understand gene expression programs during aposporous embryo sac formation. Ovules isolated from developing floral capitula of apomictic H. praealtum at capitulum stages 3 and 4 were fixed, embedded, and serially sectioned. These developmental stages contain a high frequency of enlarging AI cells with an undivided nucleus and EAE sacs containing two to four nuclei developing near a megaspore tetrad and/or degenerating megaspores (Fig. 1B; Koltunow et al., 1998, 2000). AI cells with undivided nuclei (Supplemental Fig. S1, A–C) and EAE sacs containing two to four nuclei were captured as independent samples, in addition to randomly harvested groups of SO cells spatially removed from cells undergoing either sexual or apomictic events. These three cell types provided RNA for comparative transcriptional analyses (Fig. 1B).
RNA purified from the three cell types was subjected to two rounds of linear amplification (Supplemental Fig. S1, D and E) to produce an aRNA stock free from genomic DNA (Supplemental Fig. S1F; Supplemental Table S1). The quality of the aRNA was examined by PCR for the potential to detect 100 low-copy, ovary-enriched transcripts identified in apomictic H. praealtum and H. piloselloides, which both possess the LOA-carrying chromosome (Koltunow et al., 2011a; Okada et al., 2011). The majority of these low-copy transcripts were randomly selected from a set of clones derived from cold plaque screening of an H. piloselloides ovule complementary DNA (cDNA) library (Tucker et al., 2001; see “Materials and Methods”). The remainder were genes with known spatial expression patterns in sexual and apomictic Hieracium spp. ovules (Supplemental Table S2).
Even though H. piloselloides-derived primers were primarily used in reverse transcription (RT)-PCR analyses, 75 of the low-copy ovule genes were detected in aRNA from one or more of the three H. praealtum ovule laser-captured cell types (Fig. 1C; Supplemental Table S2), suggesting strong representation of low-copy sequences in the recovered aRNA. Thirty-five of the 75 genes were expressed in all three captured cell types (Fig. 1C, class I), including Hieracium spp. FERTILIZATION INDEPENDENT ENDOSPERM, previously shown by in situ hybridization to be expressed throughout the Hieracium spp. ovule during reproductive development (Rodrigues et al., 2008; Supplemental Fig. S1G). The remaining 40 genes were differentially expressed in the three cell types (Fig. 1C, classes II–VII). Three of these, showing homology to an abscisic acid-induced RESPONSIVE TO DESSICATION22 (RD22)-like gene (clone 9.45; Supplemental Table S2), a COILED-COIL NUCLEOTIDE BINDING SITE LEUCINE RICH REPEAT (CC-NBS-LRR)-like disease resistance gene (clone 24.04; Supplemental Table S2), and a putative LIPOXYGENASE (LOX)-like gene (clone 27.18; Supplemental Table S2), were observed to be up-regulated in the AI cell by RT-PCR (Fig. 1C, class II). When quantitative real-time PCR (Q-PCR) was used to examine expression of the three AI cell-expressed genes and three others in the aRNA samples, expression patterns were consistent with the cell type enrichment patterns observed in RT-PCR, with low-level expression of the three AI cell-enriched genes detected in SO cell and EAE sac samples (Supplemental Fig. S2).
Spatial expression of the three AI cell-expressed genes was examined by in situ hybridization in apomictic H. praealtum and sexual H. pilosella during early ovule development. Figure 1, D to G, shows the expression pattern for the LOX gene in H. praealtum. The expression patterns of the other two genes are shown in Supplemental Figure S3. Transcripts were difficult to detect because of low expression levels. However, transcripts were found in enlarged, uninucleate AI cells, degenerating megaspores, degenerating nucellar epidermal cells, and EAE sacs. By contrast, transcripts from these three genes were not detectable by in situ hybridization in sexual H. pilosella ovules undergoing the events of megasporogenesis. This suggests that these three genes are up-regulated in a small subset of ovule cell types undergoing apomixis initiation and sexual suppression in the apomict.
Because the aRNA generated from the three H. praealtum laser-captured ovule cell types retained a majority of the tested low-copy ovule genes, the samples were further processed for 454 pyrosequencing to compare expression profiles in each cell type and to explore the functional identity of the AI cell. The identified set of low-level Hieracium spp. ovule sequences with known expression patterns in the three aRNA cell types served as useful internal controls to gauge the efficacy of transcript sequencing depth and assembly.
De Novo Assembly and Annotation of Expressed Sequences in Three Ovule Cell Types
Hieracium subgenus Pilosella species currently lack a reference genome or substantial DNA or EST public sequence resources, and therefore expression profiling requires the use of de novo transcriptome assembly and characterization approaches. Relative to other high-throughput sequencing technologies, such as Illumina sequencing, 454 pyrosequencing technology generates sequence reads on average 2.5 times longer, which better facilitates de novo assembly; however, the total read count is lower. In total, 465,191 high-quality sequence reads with a median read length of 251 bases were obtained from the AI, SO cells, and EAE sac samples (Table I). A de novo transcriptome characterization strategy, encompassing three complementary in silico approaches (Supplemental Fig. S4), was used to make qualitative comparisons between the cell type transcriptomes and to infer distinctive functional features of the three H. praealtum ovule cell types. The first in silico approach explored the sequence overlap between the three cell type transcriptomes, irrespective of similarity to known genes or functional annotations. This approach required the assembly of sequence reads into cell type contig sets as described below. The second and third approaches mapped expressed sequences or contigs to known protein domain and gene annotations and contrasted cell type transcriptomes in pairwise comparisons of these annotations.
Table I. Summary of RNA sequencing reads generated from apomictic H. praealtum laser-captured ovule cell types and their annotation with known protein domains.
| SO Cells | AI Cells | EAE Sacs | |
|---|---|---|---|
| High-quality reads | 167,628 | 183,420 | 114,503 |
| Median read length | 264 | 260 | 216 |
| Sequences unique to cella | 61,652 | 68,692 | 42,516 |
| Reads with Pfam matchb | 38,667 (23.1%) | 42,962 (23.4%) | 17,924 (15.6%) |
| Unique Pfam domains matched | 1,570 | 1,552 | 981 |
Sequences were deemed nonunique if sequence alignment with BLASTn (E value threshold < 1E–10) found a minimum 80% overlap with a sequence in another cell type. bSequences aligned to the Pfam protein families database (version 26.0; Punta et al., 2012) with BLASTx (E value threshold < 1E–5).
To compare the expressed sequence complement of three cell types, four high-quality sequence data sets were independently assembled using the Mimicking Intelligent Read Assembly algorithm (Chevreux et al., 2004; Supplemental Fig. S4), one for each cell type, in addition to a combined set of all three cell type data sets (Table II). A total of 8,044 sequence contigs were assembled for captured SO cells, 8,780 for AI cells, 5,002 for EAE sacs, and 18,219 for the combined assembly with median lengths of 403 to 474 bases across all four assemblies. The number of contigs generated in the combined assembly resulted in a total number of distinct contigs 16% less than the sum of the three cell type contig sets, with little gain in median contig length. This result may suggest substantial sequence diversity in these polyploid transcriptomes but may also indicate insufficient continuity of coverage across transcript lengths to achieve longer transcript assembly. The set of nonredundant contigs arising from the combined assembly was used as a consensus or point of reference for sequence comparison between the three cell type contig sets (Fig. 2A).
Table II. Summary of contigs assembled from RNA sequencing of three apomictic H. praealtum ovule cell types and a combined aggregate of their reads.
| SO Cells | AI Cells | EAE Sacs | Combined | |
|---|---|---|---|---|
| Total contig no. | 8,044 | 8,780 | 5,002 | 18,219 |
| Median contig length (bp) | 474 | 453 | 403 | 487 |
| Hieracium spp. low ovary-expressed genes observeda | 55/69 | 44/54 | 40/53 | |
| Correlation with Q-PCRb | 0.96 ± 0.05 | 0.92 ± 0.12 | 0.98 ± 0.03 | |
| Combined contigs matched in each cell typec | 6,299 | 6,778 | 3,989 | 17,066 |
| AI contigs matched in other cell typesc | 3,291 | 2,557 | 6,778 |
Number of low-expressed ovary-enriched genes found in assembled contigs is divided by the number of genes confirmed by RT-PCR analysis found in aRNA used for 454 sequencing (Fig. 1C; Supplemental Table S2). bCorrelation of the number of normalized sequence reads in assembled contigs and their expression in the three LCM samples by Q-PCR. Five randomly chosen contigs were compared for each cell type and Pearson correlation with sd is shown. cContig sequences were considered matched if reaching a BLASTn E value threshold less than 1E–10 and minimum of 80% overlap.
Figure 2.
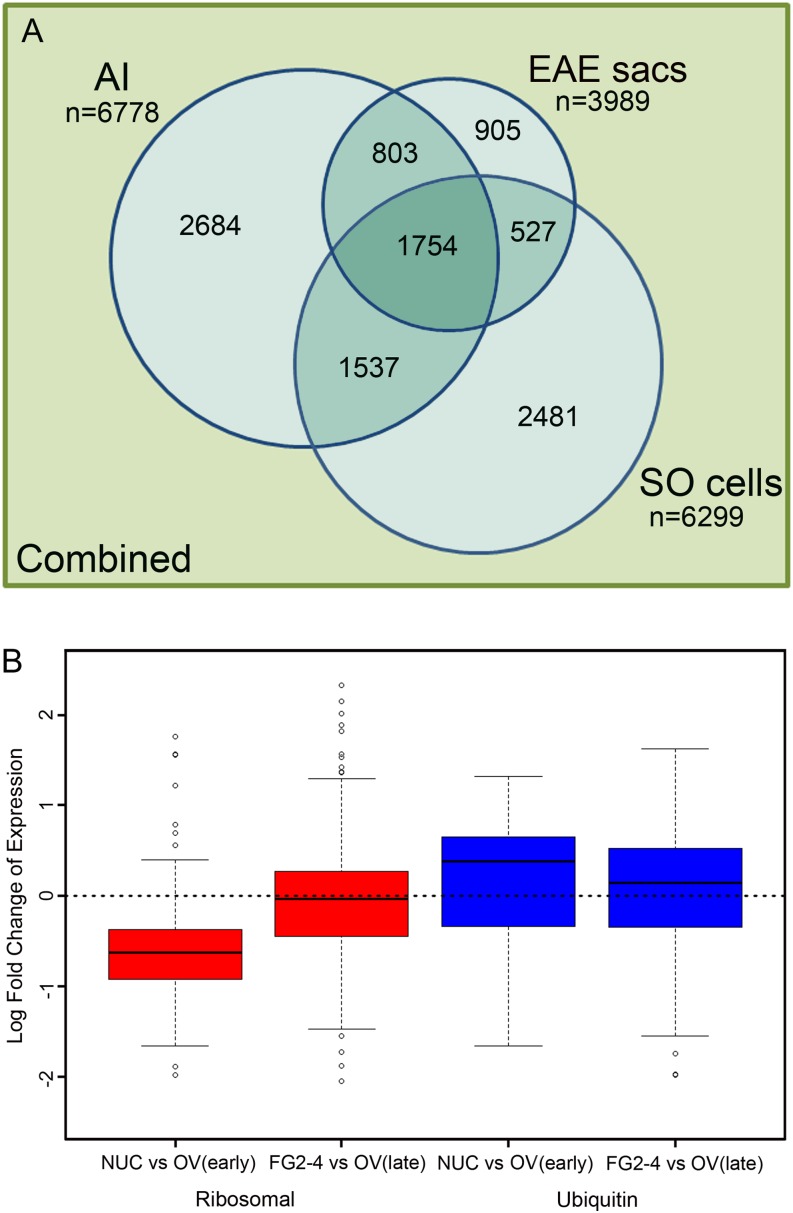
Hieracium spp. ovule cell type contig sequence comparisons and analysis of ribosomal and ubiquitin-associated gene expression in Arabidopsis ovules. A, Venn diagram showing numbers of contigs from the combined Hieracium spp. cell type assembly aligning to the cell type transcriptome contigs. Matching contigs were identified through pairwise sequence comparison as those that met a BLASTn threshold (E value ≤ 1E–10) for at least 80% of the length of the shorter contig. Where equally high-scoring or redundant matches were found, a single match was counted. Contig sequences were compared with databases with annotation and pairwise GO analyses. Statistics for these analyses are presented in Table III. B, Box plots of ribosomal and ubiquitin-associated gene expression in sexual Arabidopsis in nucellar ovule tissues compared with developing female gametophytes.
The utility and relevance of the assembled contig sets were assessed by comparison to characterized sequences expressed in the H. praealtum ovary and through annotation by sequence homology to public sequence databases. Of the low-copy ovule sequences detected by RT-PCR in RNA amplified from each cell type (Fig. 1C; Supplemental Table S2), between 75% to 79% of sequences were identified in the assembled cell type contigs (Table II). The three genes shown by in situ to be enriched in AI cells, degenerating megaspores, and nucellar epidermal cells (Fig. 1, D–G; Supplemental Fig. S3) were not detected in the assembled AI cell transcriptome. This may relate to the depth of the 454-sequence data set, such that these transcripts are below the limit of detection. In addition, the enrichment of 3′-end sequences in our transcriptome due to the RNA amplification process as found by others (Wuest et al., 2010) may hamper the identification of these sequences in the sequence read data if the characterized transcript sequences are not full length.
The number of unique contig sequences observed for the EAE sac sample was approximately 30% less than both AI cell and SO cell samples (Table II). Despite this, the ovary-enriched, low-abundance transcripts were observed in similar proportions across all cell types, including the EAE sac contig set (Table II). These observations suggest that the three LCM transcriptome data sets have comparable sequence coverage. However, until a comprehensive de novo characterization of these cell-specific transcriptomes is made possible with deeper sequence coverage, a difference in transcriptome diversity and coverage cannot be excluded. Lack of sequence depth in this data set enforces a technical boundary on the transcripts that can be observed. However, the detection of 75% of the tested low-abundance ovule transcripts in the aRNA sample and the presence of the majority of these in the assembled contig sequences suggest that the data set has a range of detection that can be used to make qualitative comparisons between these previously unstudied cell type transcriptomes.
The majority of contigs generated high-quality matches through BLAST sequence alignments to the National Center for Biotechnology Information (NCBI) nonredundant proteins, The Arabidopsis Information Resource 10 (TAIR10) peptide, and the Compositae EST databases (Table III; Supplemental Fig. S4). Hieracium species, like other Compositae, are found in the Asterids, while Arabidopsis is a member of the Rosids. High-scoring matches to both the Arabidopsis TAIR10 or Compositae sequence databases were generated for 30% to 50% sequence contigs across the cell types, and of the TAIR10 alignments, more than 96% of annotated contigs (36% of all contigs) could be mapped to a Gene Ontology (GO) term (Table III). The unannotated remainder may contain novel transcripts unique to Hieracium spp., incorrectly assembled contigs, or contigs lacking sufficiently long stretches of coding sequence to derive high-scoring cross-genome alignments.
Table III. Annotation of contig sequences assembled from apomictic H. praealtum ovule cell RNA and the expression profiles of corresponding Arabidopsis homologs in Arabidopsis ovules.
| SO Cells | AI Cells | EAE Sacs | |
|---|---|---|---|
| Total contig no. | 8,044 (100%) | 8,780 (100%) | 5,002 (100%) |
| Contigs with NCBI nonredundant protein matcha | 6,259 (77.8%) | 6,481 (73.8%) | 3,396 (67.9%) |
| Contigs with Compositae EST matchb | 3,071 (38.2%) | 3,050 (34.7%) | 1,656 (33.1%) |
| Contigs with Arabidopsis peptide matchc | 4,068 (50.6%) | 3,953 (45.0%) | 1,893 (37.8%) |
| Contigs with Arabidopsis GO annotation | 4,032 (50.1%) | 3,896 (44.4%) | 1,820 (36.3%) |
| Unique Arabidopsis array probes matchedd | 7,420 (100%) | 6,706 (100%) | 3,911 (100%) |
| Expressed in Arabidopsis nucellus or ovule at meiosis | 6,673 (89.9%) | 6,032 (89.9%) | 3,514 (89.8%) |
| Differentially expressed between Arabidopsis nucellus and ovule at meiosis | 259 (3.5%) | 210 (3.1%) | 160 (4.1%) |
| Expressed in Arabidopsis female gametophyte or postmeiotic ovule (FG2-4) | 6,084 (82.0%) | 5,746 (86.4%) | 3,249 (83.1%) |
| Differentially expressed between Arabidopsis female gametophyte and postmeiotic ovule (FG2-4) | 1,004 (13.5%) | 961 (14.3%) | 547 (14.0%) |
Contigs with a BLASTx E value threshold less than 1E–5 to the NCBI nonredundant protein database. bContigs with a BLASTn E value threshold less than 1E–10 with a minimum sequence overlap of 80% to the Compositae Genome Project EST collection (http://compgenomics.ucdavis.edu). cContigs with a BLASTx E value threshold less than 1E–5 with a minimum sequence overlap of 80% to Arabidopsis (TAIR10). dUnique Arabidopsis Agilent 4x44K array probes targeting Arabidopsis homologs of H. praealtum RNA contigs assembled from each cell type.
Although the aim of this study was qualitative comparison and de novo characterization and not quantitative profiling of transcripts, sequence read number per contig was compared with estimates of transcript abundance generated by Q-PCR for 15 randomly selected contig sequences, five contigs from each of the three cell type assemblies. For these tested candidates, the average correlation between read counts and Q-PCR for each cell type was high (R ≥ 0.9; Table II; Supplemental Fig. S5).
Collectively, these results suggest that the assembled contig sets generated from 454 sequencing of apomictic H. praealtum LCM-derived samples contain cell type-specific sequences corresponding to known transcripts from other databases, in addition to unknown and unannotated sequences.
Sequence Similarity and Overlap in Pairwise Comparisons of Cell Type Contigs
The first in silico approach explored sequence similarity between the three assembled ovule cell type transcriptomes using the total combined contig set (Table II) as a point of reference. Sequence similarity was identified through BLASTn analysis (E value ≤ 1E–10) across at least 80% of the smaller contig length. The results are presented as a Venn diagram (Fig. 2A), where the numbers represent the unique contigs from the combined contig assembly (total 18,219) observed in each cell type (Table II). In this approach, 93.7% (17,066) of the combined contig set (Table II) matched at least one contig in any cell type contig set (Fig. 2A), and 1,754 contigs were shared across all three cell types.
Approximately 40% (2,684) of all AI cell-matched contigs (6,778) were specific to the AI cell (Fig. 2A). In this respect, proportions of contigs uniquely observed in EAE sacs and SO cells were 22.7% (905) and 39.4% (2,481), respectively (Fig. 2A). Of the AI cell contigs also seen in other cell types, 22.7% (1,537) of these were specific to the AI and SO cell overlap compared with 11.8% (803) that were specific to the AI and EAE sac overlap. SO cells also shared more contigs uniquely with AI cells (24.4%) than with EAE sacs (8.4%). From the perspective of the EAE sac transcriptome, there were slightly more contigs unique to the overlap between EAE sacs and AI cells (803) than there were unique to the overlap between EAE sacs and SO cells (527; Fig. 2A). Thus, in terms of similarity of expressed sequences, the EAE sac transcriptome bore greatest similarity to the AI cell, while the AI and SO cell transcriptomes shared greater sequence overlap than either of their pairwise sequence comparisons with EAE sacs.
Protein Domain Annotations of Unassembled Sequences Show Cell Type-Specific Enrichment in Signaling and Protein Metabolism
The second in silico approach investigated each cell type for specific protein domain signatures that may reflect distinctive functional attributes. We analyzed the set of high-quality unassembled sequences from each cell type using Pfam, the protein domain sequence database resource (Punta et al., 2012). Reads from the SO cells, AI cell, and EAE sac samples could be mapped to 1,570, 1,552, and 981 Pfam domains, respectively (Table I). Significantly enriched Pfam domains for each cell type were identified in pairwise contrasts between the three cell types (Supplemental Fig. S4) and annotated for GO categories. Supplemental Table S3 shows the set of significantly enriched Pfam domain annotations in each cell type with associated GO terms.
In the context of enriched Pfam domains with GO annotations, the greatest distinctions could be found in comparisons between EAE sacs and SO cells, with a total of 23 Pfam domains showing differential frequencies between these two cell types. Of the 15 domains that show enrichment in EAE sacs, nine represent either small or large subunits of the ribosomal complex (Supplemental Table S3), and as such, all are associated with the GO molecular function term of structural constituent of ribosome (GO:0003735). Most other domains found enriched in the EAE sac data relative to SO could be grouped under the GO parent terms of hydrolase activity (GO:0016787) or transition metal ion binding (GO:0046914), with the latter enriched in cytochrome P450 (iron binding) and plastocyanin-like (copper ion binding) domains. Each of these domains has been identified as an important catalytic component in a broad range of physiological, developmental, and signaling pathways in plants.
Comparison of the AI cell with SO cells showed enrichment of ribosomal protein Pfam domains in the AI cell, similar to that found in the EAE sacs relative to SO cells. However, the AI cell was also enriched for Pfam domains implicated in ubiquitin-dependent protein degradation. The ubiquitin proteasome protein catabolic complex is involved in diverse developmental processes, including regulation of auxin (for review, see Vierstra, 2012) and jasmonate signaling (Xie et al., 1998), pathogen resistance (Austin et al., 2002), cell cycle progression, and gametophyte development (Honys and Twell, 2004; O’Brien et al., 2004; Liu et al., 2008; Juranič et al., 2012). The S-phase Kinase-Associated Protein 1 (SKP1) Cullin F-Box protein E3 ligase complex marks target proteins for degradation via ATP-dependent covalent attachment of ubiquitin. Enrichment of SKP1 proteins in the AI cell was represented by enrichment of the POZ domain (Skp1_POZ) found at the N terminus and the SKP1 dimerization domain found at the C terminus of the protein (Supplemental Table S3).
The AI cell and EAE sac Pfam domain comparisons showed the least differences, implying a greater functional similarity. The majority of domains observed to be enriched in the SO cell sequence set relative to EAE sacs were similarly enriched in SO cells relative to the AI cell, also supporting closer functional association between AI cells and EAE sacs relative to SO cells. The only domain showing statistically significant enrichment in AI cells relative to EAE sacs was the WD40 protein domain. This domain has been implicated in female gametophyte development (Shi et al., 2005) and is known to function in creating protein scaffolds and facilitating protein interactions in multiprotein complexes such as the E3 ligase complex (Smith et al., 1999). Pectinesterase, Cys protease, and the profilin domain were enriched in EAE sacs relative to the AI cell sequence set, which may relate to events of embryo sac growth and expansion. The profilin domain is implicated in actin binding, commensurate with the abundance of actin cytoskeletons in two- and four-nucleate embryo sacs during megagametogenesis (Webb and Gunning, 1994).
Collectively, analysis of protein domains that could be annotated in unassembled reads from the three captured cell types suggested that the SO cell transcriptome was functionally distinct from those of the AI and EAE sac, predominately in signaling and ribosome- and ubiquitin-related domains, while highlighting substantial functional overlap between the AI cell and dividing EAE sacs.
Annotation of Assembled Contigs Shows Closest Functional Identity Exists between AI Cells and EAE Sacs
In a third in silico approach, we associated contigs to GO annotations through sequence homology and sought to identify GO terms that showed specific cell type enrichment using pairwise comparisons. All contig sets showed comparable GO annotation rates ranging from 65% to 74% (Table III). GO enrichment analysis was completed in two complementary stages; the first was a commonly used singular GO term enrichment analysis (SEA) utilizing a Fisher exact test for increased frequencies of individual terms in an input list relative to a background list (Ashburner et al., 2000; Du et al., 2010). While a commonly used and powerful approach, SEA GO analyses consider individual GO term frequencies in isolation against the full GO hierarchy and therefore can struggle to identify statistically significant evidence of enrichment for more specific, but less frequently observed, child terms. To address this, nested GO analysis using nested Expression Analysis Systematic Explorer (nEASE) was also used (Chittenden et al., 2012). This approach restricts testing to within significant terms found in the first stage (SEA) and uses related GO terms and similarly annotated genes to better discriminate enrichments at more specific GO terms. These approaches have been shown to provide greater sensitivity to detect biologically relevant functional themes in human cancer expression profiling (Zhang et al., 2010; Chittenden et al., 2012). The full results of the GO enrichment analysis from both singular and nested analyses are presented in Supplemental Table S4.
Interestingly, there were no statistically significant (P ≤ 0.05) differences in GO term counts identified in the reciprocal pairwise comparison between AI cell and EAE sac derived annotations. This implied close functional identity between these two cell types (Supplemental Table S4) but does not necessarily mean that the same genes are expressed in both samples, and clearly all cell type contig sets contain subsets of unique expressed sequences (Fig. 2A). The subsets of contig sequences unique to AI and EAE sacs were further explored, and annotation rates and GO results were concordant with comparisons of the full cell type contig sets. Therefore, these unique sequences may represent different sequence mappings to similar gene sets, poorly annotated sequences, or species-specific sequences.
The pairwise comparison of EAE sac and SO cell contig annotations yielded the greatest number of discriminatory GO terms (Supplemental Table S4). These findings were congruent with the prior Pfam annotation analysis of unassembled sequence reads. The enriched GO annotations of EAE sac contigs relative to SO cells are dominated by terms related to gametophyte development, lipid localization, ribosome biogenesis, translation, and gene expression, overlapping with many of the annotated Pfam domains. The functional terms enriched in AI cells relative to SO cells also shared substantial overlap with those enriched in EAE sacs relative to SO cells; however, the biological process of flower development was uniquely enriched in AI cells relative to SO cells (Supplemental Table S4).
GO enrichment terms in SO cells relative to EAE sacs centered on functional themes of signaling, protein kinase activity and phosphotransferase activity, transcription factors, and nucleic acid metabolism (Supplemental Table S4). Similar functional themes were enriched in SO cells relative to the AI cell annotations, along with increased annotations related to glycosyl hydrolysis and methyltransferase.
In summary, the pairwise comparisons between assembled contig sequences from each cell type, made on the basis of sequence similarity alone, indicated that while the EAE sac-expressed contigs shared greater similarity to the AI cell than SO-expressed contigs, the AI cell shared best overlap with SO cell-expressed contigs. In complementary annotation-based analyses that identify functional similarity (derived GO annotations), very few discriminatory GO terms from either gene or Pfam domain annotations were identified between AI cell and EAE sac. Derived GO annotations from contigs and Pfam domain annotations of unassembled reads both suggest relative enrichment of signaling pathways in SO cells and overlapping enrichment of ribosome biogenesis, translation, and gametophyte development in the AI and EAE sac transcriptomes. The Pfam analysis more clearly proposes an enrichment of ubiquitin pathway-associated expression in AI and EAE sacs than the contig analysis. Conversely, the contig analyses propose more extensive and specific enrichment of terms in signaling pathways associated with the SO cell transcriptome. This analysis suggests that the AI cell transcriptome shares molecular signatures with both the SO cell and EAE sac transcriptomes but is functionally more similar to the EAE sac and is consistent with the AI cell undergoing a developmental transition between the SO cell type and the EAE sac.
Expressed Genes in Apomictic H. praealtum Ovules Share Homologs with Arabidopsis Genes Expressed Early in Female Gametophyte Development
We investigated the possibility that the apomictic H. praealtum ovule transcriptome assemblies contained transcripts of general relevance to sexual ovule reproductive cell types and whether temporal expression patterns were shared with cell-specific expression profiles of comparable stages of sexual Arabidopsis female gametophyte development. The Arabidopsis LCM expression data sets analyzed were generated with the Agilent 4x44K Arabidopsis array and captured gene expression from the nucellus at meiosis, the whole ovule at meiosis, the postmeiotic female gametophyte at megagametogenesis containing two to four nuclei (FG2-4), and the whole ovule at FG2-4 stage (Tucker et al., 2012a).
The Hieracium spp. cell type contigs were mapped to Arabidopsis gene peptide sequences (TAIR10) using BLASTx, and numbers of matches ranged from 1,893 (38%) to 4,068 (50%) across the cell type assemblies (Table III). This corresponded to a range of between 3,911 and 7,420 unique Arabidopsis gene expression probes on the Agilent array (44K, Table III). The Hieracium spp. contig sets mapped to the array were found to be dominated by Arabidopsis ovule-expressed sequences (83%–90%; Table III). After filtering to remove genes present in all seven data sets, radar plots were used to examine the similarity between the three Hieracium spp. and four Arabidopsis data sets (Supplemental Fig. S6). Interestingly, no obvious trends were detected to suggest that, based on Arabidopsis gene identity, the Hieracium spp. cell type contigs were biased for expression in any one Arabidopsis cell type.
Genes that were differentially expressed between tissue types in Arabidopsis and could be mapped to an H. praealtum ovule contig were also examined (Table III; Supplemental Fig. S7). Genes meeting these criteria were more likely to be differentially expressed between the FG2-4 and whole ovule sample, compared with the nucellus and its corresponding whole ovule sample (Table III; Supplemental Fig. S7). Many of these genes encoded ribosomal subunits. Analyses of the expression of ribosomal genes in Arabidopsis ovules showed that most ribosomal genes are decreased in abundance in the Arabidopsis nucellus relative to other tissues, but ribosomes become more abundant in the female gametophyte at FG2-4 during early mitotic events (Fig. 2B). Thus, the enrichment of ribosomal genes in cell types undergoing female gametophyte development in sexual Arabidopsis parallels the enrichment of ribosomal genes in AI cells and EAE sacs in apomictic H. praealtum relative to other ovule cell types during EAE sac development.
We next queried the expression of ubiquitin-associated genes in the Arabidopsis nucellus relative to other Arabidopsis ovule tissues. Figure 2B shows that ubiquitin-associated genes are slightly more enriched in the nucellus compared with other ovule tissues (Fig. 2B). Reciprocal best BLAST analyses between the 329 probes associated with ubiquitin processes on the Arabidopsis array and Hieracium spp. AI cell, SO cell, and EAE sac contigs show that 16 are found only in the AI cell relative to eight in the SO cell and four in EAE sacs. Genes expressed in the AI cell-only category were RELATED TO UBIQUITIN1 (RUB1) and SKP1-like and included some ubiquitin protein ligases, which is consistent with the enriched ubiquitin- associated domains found in the Pfam analyses (Supplemental Table S3). However, in apomictic H. praealtum, ubiquitin-associated sequences are enriched in both AI cells and EAE sacs (Supplemental Table S3; Fig. 3).
Figure 3.
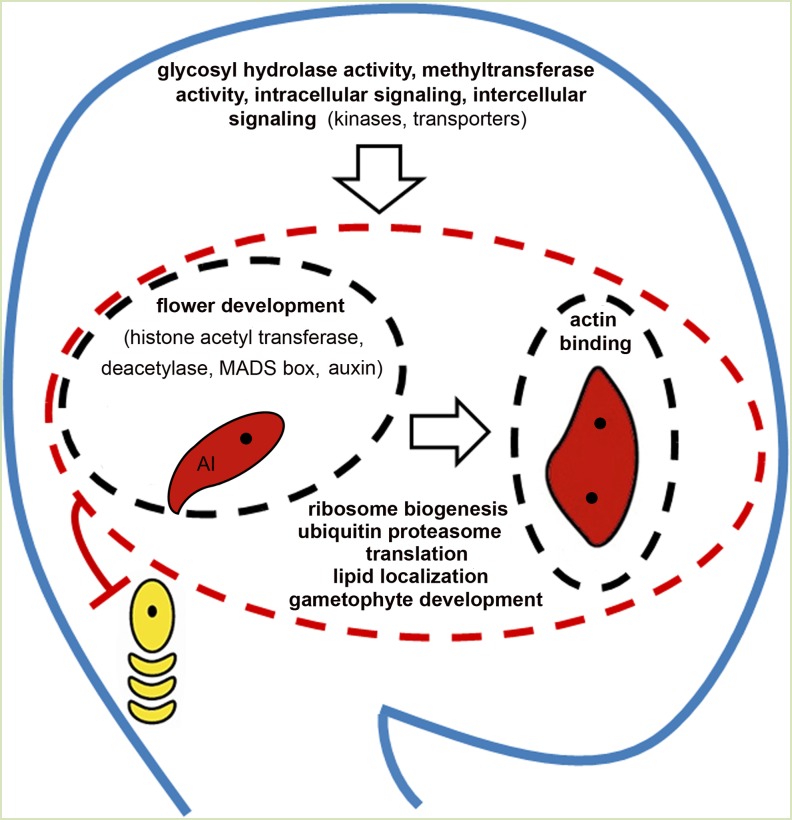
Summary of enriched gene functional categories in the three apomictic ovule cell types from Pfam and assembled contig analyses. The uninucleate AI cell and EAE sac containing two to four nuclei (shaded red) show preferential enrichment of functional terms shown inside the black dashed ovals surrounding them. They share many functional terms indicated within the red dashed oval surrounding both cell types. The remaining terms in the ovule cartoon relate to significant functional categories found in the SO cells. This complement of functional categories results in the demise of the adjacent sexually derived haploid megaspores (yellow).
Taken together, these comparisons of gene expression in ovules of sexual Arabidopsis and apomictic H. praealtum at comparable stages of gametophyte development show high similarity in sequence identity of expressed genes. However, their differential behavior in the tissue sets is not absolutely conserved. This may be due to a combination of differences in the evolutionary distance between the two species, ploidy of the embryo sac structures (i.e. the diploid H. praealtum EAE sac versus the haploid meiotically derived Arabidopsis female gametophyte), differences in the developmental stage of the tissues collected, and possibly heterochronic ovule gene expression relating to aposporous embryo sac growth.
Meiosis and Megaspore Gene Homologs Are Not Found in Enlarging AI Cells
We utilized the significant annotation overlaps between Arabidopsis and Hieracium spp. to directly query data sets for putative homologs in each captured sample to further investigate indicative AI cell functions. First, we queried whether the AI transcriptome bears similarity to that of the MMC or FM (Fig. 1A). Annotation of the AI cell and EAE sac contigs did not yield any GO annotations related to meiosis function. We directly queried the contig set and the unassembled read set for currently known genes characteristic of and/or required for function of these meiotic and postmeiotic cell types. Transcripts resembling meiosis genes that are highly conserved in plants and animals, such as DMC1, SPORULATION11, RAD50, ASYNAPTIC1, and MUTS HOMOLOG4 (Schwarzacher, 2003), were not observed in any of the cell type transcriptome sequence sets. Other genes required in meiotic cells of Arabidopsis, including SOLO DANCERS, SWITCH1/DYAD, MULTIPOLAR SPINDLE1, SPOROCYTELESS/NOZZLE, MUTL-HOMOLOG1, REC8, PARTING DANCER, and TARDY ASYNCHRONOUS MEIOSIS, were similarly not detected. Furthermore, homologs of Arabidopsis FM genes, CHROMATIN-REMODELING PROTEIN11, FM, MNEME, MtN3, and ARABINOGALACTAN PROTEIN18 gene transcripts were not observed in the AI cell transcriptome (see Supplemental Table S5 for Arabidopsis gene identifiers). As sequence coverage in this study is not saturating, we investigated the possibility that these meiosis-associated genes are expressed in the ovule at levels below the detection threshold of the data set. Of the 19 Arabidopsis meiosis-related homologs investigated, 16 were represented by probes on the array and seven were detected with robust expression values (greater than the median of all probes) within the Arabidopsis ovule samples. As mentioned above, low-abundance transcripts are observed in the H. praealtum ovule data set. These ovule contigs were successfully mapped to many putative Arabidopsis homologs showing abundance in the Arabidopsis ovule less than the median expression value. While it is still possible that some of these transcripts are present at levels below the range of detection of expression of this data set, it would be expected that sequencing depth is sufficient to detect at least the seven conserved candidates with robust transcript abundance in the Arabidopsis ovule. The absence of any of the known transcripts required for meiosis and FM function in the AI cell suggests it is unlikely to be undergoing either a meiotic or FM developmental program.
Unique Developmental Regulators Are Expressed in Developing AI Cells as Revealed by GO Analysis
To interrogate the possible molecular functions of the AI cell further, we examined more closely the Arabidopsis genes identified by sequence homology found to be differentially present in the three ovule cell types. To guide this investigation, we focused on genes associated with the statistically significant GO terms uncovered by the nEASE in pairwise comparisons involving the AI cell (Supplemental Table S4). This analysis had identified six GO terms in the AI and SO cell pairwise comparison and none in the AI and EAE sac comparison (Supplemental Table S4). SO-enriched terms included carbohydrate metabolic process and methyltransferase activity, while AI-enriched terms included flower development, gametophyte development, multi-organism process, and lipid localization. Notably, lipid localization, multi-organism process, and gametophyte development was also enriched in EAE sacs relative to SO cell by nEASE; Supplemental Table S4). The list of genes underlying these terms was filtered to remove the genes identified in both AI and SO cell transcriptomes, leaving the annotations specific to the AI, EAE sacs, or SO cell type. These annotations were also queried for evidence of expression in Arabidopsis early ovule tissues as assessed by the 44K array (Supplemental Table S6; Tucker et al., 2012a).
The AI cell exhibits enriched expression of putative homologs of Arabidopsis histones, histone acetyl, and deacetyl transferases and floral genes involved in chromatin function and gene silencing via small RNA pathways (DICER2-like; Supplemental Table S6). Members of chromatin complexes regulating expression of auxin-related genes, meristem maintenance, and auxin efflux were observed. A putative homolog of UNFERTILIZED EMBRYO SAC5 (UNE5), a stress-induced protein thought to be involved in secretory pathways, was also observed. Mutations in UNE5 in Arabidopsis lead to defective fertilization in embryo sacs (Supplemental Table S6; Pagnussat et al., 2005). Rho GTPase-like genes were found, and these act as molecular switches that control cytoskeletal dynamics and influence pollen tube tip growth and animal cell movement in spatial cell zones. Their expression may suggest potential involvement in the directional growth of AI cells toward sexual cells during sexual suppression (Kenneth and Duckett, 2012; Supplemental Table S6).
Both AI cells and EAE sacs are enriched in expression of genes resembling those involved in the cell cycle G-to-M transition (CELL DIVISION CYCLE5-like) or found in root and shoot meristematic regions (TRANSLATIONALLY CONTROLLED TUMOR PROTEIN-like). A homolog of EMBRYO SAC DEVELOPMENTAL ARREST27 (EDA27) was enriched in both cell types. Mutations in EDA27 lead to unfused polar nuclei. Interestingly, a putative homolog of the basic helix-loop-helix BEL1-like gene EOSTRE (AT2G35940; Supplemental Table S6) was detected in AI cells and two to four nucleate EAE sacs. Transcripts of this gene are not detected during mitosis in Arabidopsis embryo sacs. Ectopic expression of EOSTRE results in abnormal nuclear migrations, and one of the synergids is converted to a second egg cell in 10% to 15% of cases (Pagnussat et al., 2007). Embryo sac formation in apomictic Hieracium spp. also exhibits abnormal nuclear migration and conversion of synergids to eggs, leading to corresponding frequencies of polyembryonic seed formation (Koltunow et al., 1998, 2000). This may reflect a misexpression of a similar gene during apospory; however, expression comparisons with sexual Hieracium spp. embryo sacs are needed to confirm this.
In general, this expanded in silico analysis of genes underlying terms found through nested GO enrichment highlights the similarity in mitotic embryo sac-like programs between AI cells, EAE sacs, and Arabidopsis embryo sacs (Supplemental Table S6) and reveals potential ectopic gene expression and/or possible gene recruitments that may influence features of AI cell gene expression and fate.
Stress and Disease Resistance-Like Gene Expression in AI Cells and EAE Sacs
AI cells and EAE sacs also appear to exhibit expression of homologs of genes not evident in Arabidopsis embryo sacs at two to four nucleate stages from array analyses (Supplemental Table S6). These include abscisic acid stress-inducible genes thought to function in autophagy, a CONSTITUTIVE DISEASE RESISTANCE1-like homolog, and pathogenesis-associated lipid transfer-like proteins (Supplemental Table S6). Homologs of other genes involved in responses to salt, ozone, other abiotic stresses, and pathogen infection were evident in AI cells and/or EAE sacs (Supplemental Table S6). The significance of this is unclear and may reflect involvement in aspects of H. praealtum aposporous embryo sac growth. However, homologs of three genes that fit into this “stress-pathogen” category include an abscisic acid-induced RD22-like gene, a CC-NBS-LRR-like resistance gene, and a putative LOX gene coordinately up-regulated in the AI cell, degenerating nucellar epidermal cells and megaspores in apomictic ovules. These are undetectable by in situ in sexual ovules, suggesting association with apomictic initiation and sexual suppression (Fig. 1, D–G; Supplemental Fig. S3). The functional associations of such stress and pathogenesis-related genes in apomictic events warrant further investigation.
DISCUSSION
Apomixis Control and Transcriptional Analyses of Apomixis Events
Reproductive events in ovules are controlled by cell-autonomous and nonautonomous pathways (Drews and Koltunow, 2011). Apomixis has evolved many times in flowering plants (Carman, 1997), and most of the suites of observed mechanisms are controlled by dominant genetic loci. Apomixis is proposed to be manifested by recruiting the sexual machinery in a heterochronic manner so that the events of meiosis and fertilization are bypassed (Koltunow and Grossniklaus, 2003; Sharbel et al., 2010; Koltunow et al., 2011a; Tucker et al., 2012b). Genetic and epigenetic mechanisms have been proposed to regulate apomixis, with the apomixis loci postulated to influence global and cell type-specific processes during the chronology of reproductive events leading to seed formation (Koltunow and Grossniklaus, 2003; Catanach et al., 2006; Okada et al., 2011; Rodriguez-Leal and Vielle-Calzada, 2012).
To date, approaches aimed at understanding gene expression in apomicts, whose genomes currently remain unsequenced, have involved the use of RNA extracted from florets or microdissected ovules and analyses involving differential screening and comparative cDNA sequencing (Vielle-Calzada et al., 1996; Tucker et al., 2001; Rodrigues et al., 2003; Albertini et al., 2004; Singh et al., 2007; Cervigni et al., 2008; Laspina et al., 2008). More recently, high-throughput RNA sequencing technologies have been employed (Sharbel et al., 2009, 2010). Deep-sequencing analyses in microdissected diploid sexual and apomictic Boechera spp. ovules, which undergo diplospory, have identified a down-regulation of gene expression in apomictic ovules relative to sexual ovules around the time of MMC development and its switch to mitotic embryo sac formation. However, there was no obvious developmental pathway or timing change that could simply explain the shift to apomixis. Transcription factors were overrepresented among apomixis-specific genes, suggestive of large-scale regulatory changes in apomictic ovules (Hofmann, 2010; Sharbel et al., 2010).
Depending on the aposporous species, timing of AI cell development may occur at various stages in relation to the temporal sequence of sexual events. In some species, the sexual pathway persists even though aposporous embryo sacs form (Koltunow and Grossniklaus, 2003). We were unable to find obvious commonality of gene expression categories in our apomictic H. praealtum ovule cell transcriptomes compared with the available transcriptome information from aposporous grass species Poa pratensis, Paspalum notatum, and Pennisetum glaucum (Albertini et al., 2004; Laspina et al., 2008; data not shown). This may reflect sequencing depth and associated limitations in the ability to resolve differential expression in AI cells or unreduced female gametophytes in the complex starting material employed in those studies. Apospory may also involve changes in a subset of commonly expressed genes in sporophytic and gametophytic cells whose action is reflected at the posttranscriptional or protein level. Alternatively, apospory in grass species and eudicot Hieracium spp. may result from different molecular mechanisms.
While studies in whole ovules may indicate association of candidate pathways with apomictic reproduction, the cells involved in the process form a small percentage of the total ovule cell mass, spatial validation by in situ does not feature in some studies, and functional validation is limited by the ability to transform the species. Isolation of specific cell types enables direct transcriptional comparisons (Kerk et al., 2003; Day et al., 2005). Here, we have confirmed the efficacy of LCM in combination with 454 pyrosequencing, bioinformatic analyses, and in situ hybridization to explore transcriptomes of two apomictic cell types, the AI cell and the EAE sac of H. praealtum in relation to SO cells not participating in these events. We also compared our expression data with various ovule cell types isolated by laser capture from Arabidopsis.
The Enlarging AI Cell Is Transitioning to a Mitotic Embryo Sac Program prior to Nuclear Division
While the transcriptome sequence data sets obtained from the laser-captured H. praealtum ovule cell types were not saturating for quantitative analyses, low-abundance and cell-specific transcripts and protein domains were identified in conjunction with a range of putative homologs found in ovules of other sexual species. The presence of low-abundance transcripts and overlaps with putative ovule-expressed homologs suggests that this data set can provide useful insights into discriminating cellular functions of the enlarging AI cell. GO categories enriched in each examined cell type are summarized in Figure 3. These enrichments were identified through pairwise comparison of the three cell types (AI to SO cell, AI to EAE sac, and EAE sac to SO cell) and showed that the AI cell transcriptome displays a similar functional profile to the EAE sac transcriptome.
The AI cell transcriptome does not exhibit expression of meiosis-associated genes that are conserved in many plant species. Relative to SO cells and EAE sacs, the AI cell is enriched in GO terms related to flower development, with underlying gene homologs, including histone acetyl transferases and histone deacetylases, MADS box genes, and participants in auxin signal transduction. However, both the AI cell and EAE sacs share significant overlaps in GO functional categories and protein domains that are quite distinct from those seen in SO cells. The AI cells and EAE sacs are significantly enriched in gametophyte development functions, translation, ribosome biogenesis, and ubiquitin proteasome function (Fig. 3). Both cell types express homologs of a number of genes involved in Arabidopsis female gametophyte development. Ribosome biogenesis is increased in AI cells and EAE sacs of H. praealtum and also during female gametophyte development in Arabidopsis, consistent with the requirement of ribosomal genes for progression of the gametophytic cell cycle (Shi and Yang, 2011). Developing Arabidopsis embryo sacs do not exhibit the up-regulation of ubiquitin-associated pathways evident in aposporous H. praealtum AI cell and EAE sac transcriptomes. Collectively, the absence of detectable meiosis gene expression and the significant overlap in homologs of genes involved in female megagametophyte development in the AI cell and EAE sac suggests that the enlarging AI cell is transitioning to and acquiring an embryo sac fate even though it has not yet undergone nuclear division.
Sporophytic Influences on AI Cell Differentiation and Aposporous Embryo Sac Development
Signaling from SO cells, involving both genetic and epigenetic networks, plays a major role in both promoting and restricting the number of cells with sporogenic potential that can initiate female gametophyte development in premeiotic ovules of sexual species (Armenta-Medina et al., 2011; Bencivenga et al., 2011). Disruption of pathways involving Leu-rich receptor-like kinases (RLKs) and their ligands, ARGONAUTE proteins, and other members of small RNA pathways, in addition to DNA methyltransferases in sporophytic tissues of premeiotic Arabidopsis, rice (Oryza sativa), and maize (Zea mays) ovules, can result in failure of MMC formation or the development of multiple MMC-like or megaspore-like cells (Nonomura et al., 2003, 2007; Zhao et al., 2008; Garcia-Aguilar et al., 2010; Olmedo-Monfil et al., 2010). Disruption of sporophytic ARGONAUTE function in maize can lead to a change in MMC cell fate such that it bypasses meiosis and undergoes mitosis forming a diploid embryo sac (Singh et al., 2011). Recent data suggest that integrity of small RNA pathways in SO tissues is also important for sequential progression between megasporogenesis and megagametogenesis in Arabidopsis (Tucker et al., 2012a).
SO cells in apomictic H. praealtum ovules were enriched in signaling functions involving methyltransferases, various kinases, transporters, and other intra- and inter-cellular signaling molecules compared with the AI cell and EAE sacs (Fig. 3). Thus, consistent with their predicted identity, SO cells in H. praealtum possess machinery in common with ovules of sexually reproducing plants that may function in regulating cell specification events during female gametophyte development. The effects of altering methyltransferase or RLK activity and the composition of the ARGONAUTE protein family in apomictic and sexual Hieracium spp. ovules are currently unknown. Alterations in SO pattern formation in apomictic Hieracium spp. arising from targeted cellular ablation, disruption in polar auxin transport and auxin perception (Koltunow et al., 2001; Tucker et al., 2012b), and mutation (Koltunow et al., 2011a) influence the number and position of AI cells formed and progression of aposporous embryo sac formation. AI formation within the ovule is dependent upon LOA function after the initiation of sexual meiosis (Koltunow et al., 2011a). The enriched classes of signaling molecules identified here, such as kinases that influence female gametophyte development in sexual systems, may thus influence the capacity of SO cells to respond to LOA in apomictic Hieracium spp.
Roles of Ubiquitin Proteasome Pathways in Meiotic Avoidance and Apomixis
The anaphase-promoting complex/cyclosome (APC/C) is an evolutionarily conserved E3 ubiquitin ligase critical for cell cycle progression by degrading cell cycle proteins. The enrichment of ubiquitin proteasome components in AI cells observed in this study during their growth and transition to mitotic events of embryo sac formation is in keeping with its function in mitotic cell types. The Arabidopsis protein OMISSION OF THE SECOND DIVISION1 (OSD1) is associated with and negatively regulates the APC/C. OSD1 functions in both divisions of meiosis and, interestingly, loss of its function leads to omission of the second meiotic division. In osd1cyclin A1;osd1cyclin A2 double mutants, the first and second meiotic divisions are avoided (d’Erfurth et al., 2009, 2010).
Genomic sequences tightly linked to the apospory locus in tetraploid St John’s wort (Hypericum perforatum) contain a truncated gene, HpARI, homologous to Arabidopsis ARIADNE7, which encodes a ring finger E3 ligase protein predicted to be involved in various regulatory processes related to ubiquitin-mediated protein degradation. The HpARI marker cosegregates with apospory but not autonomous embryo formation and is inherited in a dominant manner in aposporous segregants. Three intact “sexual alleles” are also present in the tetraploid apomict, and these are coexpressed with the truncated HpARI gene in a variety of plant tissues. HpARI is proposed to act in a dominant negative manner at the protein level to influence alterations in gametophyte development (Schallau et al., 2010). Cys-rich ring domains characteristic of the ARIADNE family are found in contigs from all three apomictic H. praealtum laser-captured ovule cell types and merit further characterization.
A range of gene homologs up-regulated in AI cells and EAE sacs relative to SO cells are involved in processes regulated by the ubiquitin proteasome pathway, including auxin and jasmonate signaling, flower development, pathogen resistance, abiotic stress (drought), cell cycle progression, and gametophyte development. Thus, it is conceivable that the LOA locus may influence this pathway in apomictic Hieracium spp.
CONCLUSION
LCM in this study has enabled an analysis of the indicative functions of specific ovule cell types at early stages of aposporous embryo sac formation in apomictic H. praealtum. We have determined that enlarging AI cells appear to be transitioning to a gametophytic program prior to their first nuclear division. Future examination of transcriptional profiles of laser-captured cell types from sexual, apomictic, and mutant Hieracium spp. that have lost LOA and/or LOP function, in conjunction with whole ovule transcriptome analysis from the same material using the depth afforded by the Illumina short-read platform, should provide a comprehensive, quantitative analysis of large and small RNA pathways operating during apomictic reproduction.
MATERIALS AND METHODS
Plant Materials
Apomictic Hieracium praealtum accession R35 (Catanach et al., 2006; Koltunow et al., 2011a) and tetraploid sexual Hieracium pilosella accession P36 were grown in a glasshouse under long-day conditions with average day and night temperatures of 25°C and 16°C, respectively (Koltunow et al., 2011a).
Tissue Fixation, Embedding, and LCM
Farmer’s fixative (ethanol:acetic acid [3:1] containing 1 mm dithiothreitol [DTT]) was used as a fixative for H. praealtum (R35) floral tissues (Kerk et al., 2003). Capitula were collected at stages 3 and 4 of development (Koltunow et al., 1998), vertically bisected in ice-cooled fixative, placed under vacuum for one hour at –75 kPa, and stored at 4°C overnight. Ovaries were dissected from the fixed florets in 70% (v/v) ethanol and further dehydrated to 100% (v/v) ethanol. Tissue was infiltrated at 4°C with a butyl methyl methacrylate (BMM) solution (79.5% (v/v) n-butyl methacrylate, 20% (v/v) methyl methacrylate, and 0.5% (w/v) benzoin methyl ether from ProSciTech with 1 mm DTT) and ethanol in the volume ratios 1:3, 1:1, 3:1, 1:0, and 1:0, with gentle agitation for 12 h each change (Baskin et al., 1992). Individual ovaries were placed in BEEM capsules (ProSciTech) and polymerized at –20°C under UV light (6-W lamp) for 3 to 5 d. Serial ovule sections, 5 µm thick, were cut using glass knives with a rotary microtome (Model 2055, Leica Microsystems), floated on sterile water on a Leica PEN membrane-coated microscope slide, and dried at 42°C prior to long-term storage at 4°C. BMM was removed by slide immersion in 100% (v/v) acetone for 10 min and drying at 42°C prior to LCM. Cells were dissected from sections using a Leica AS Laser Microdissection system quipped with a 63× objective at aperture 5 to 6 and a UV laser (337-nm wavelength) at intensity 40 to 46 and tracking speed 3 to 4. AI cells with undivided nuclei were captured from 270 individual 5-µm sections (Supplemental Fig. S1). A total of 100 EAE sacs containing two to four nuclei were also harvested from 5-µm sections. Clusters of approximately 50 SO cells were cut from 50 individual 5-µm ovule sections after AI cells and EAE sacs had been harvested. Harvested cells were collected in the cap of a 0.2-mL PCR tube, and the RNA was either isolated immediately or the captured cells were stored at –80°C.
RNA Isolation and Amplification from LCM Sections
Total RNA was extracted from the captured cell types using a PicoPure RNA isolation kit (Arcturus Bioscience) according to the manufacturer’s instructions including the on-column DNase treatment. The RNA isolated from the three captured samples was subjected to amplification using a MessageAmp II aRNA Kit (Ambion). This protocol employs a linear amplification procedure adapted from Eberwine et al. (1992) that has been shown to be robust to bias in amplified transcript abundances (Li et al., 2004). Yields after first-round amplification of the samples were 1,300 ng for SO cells, 646 ng for AI cells, and 513 ng for EAE sacs. Ten percent of the first-round product was used for second-round amplification, producing 189 µg, 154 µg, and 126 µg, for SO cells, AI cells, and EAE sac samples, respectively. Total RNA was also extracted from whole H. praealtum R35 ovary samples at the same stage used for LCM using the RNeasy Plant Mini Kit (Qiagen) and subjected to amplification as a control. (See Supplemental Protocol S1 for preliminary analyses of isolated and aRNA).
Isolation of Low-Prevalence Ovary Sequences from Apomict Hieracium piloselloides D18 and Expression Analysis in aRNA Samples from H. praealtum (R35)
Cold plaque screening was performed on a cDNA library made from ovaries extracted from H. piloselloides (D18) at stages 2 to 4 of capitulum development (Tucker et al., 2001). A bank of cDNA clones representing predominantly ovary-specific genes expressed at low levels were randomly selected and sequenced (Tucker et al., 2001). EST sequences of 500 bp were deposited to the NCBI database (accession nos. GO673075–GO673166). BLASTx searches of the sequenced ESTs against the Arabidopsis (Arabidopsis thaliana) TAIR10 peptide database (http://www.Arabidopsis.org/index.jsp) were carried out to provide a preliminary indication of homology. E values lower than 1E–5 were considered to represent significant similarity. Putative clone identities using the best match to the BLAST results are presented in Supplemental Table S2. Primer pairs were developed for each clone for use in RT-PCR in AI cell, EAE sac, and SO cell aRNA, and these sequences are also provided in Supplemental Table S2. Approximately 15 ng of aRNA from the AI cell, EAE sac, and SO cell samples were used as a template for cDNA synthesis using the MessageAmp II aRNA Kit (Ambion). RT-PCR was performed using equal amounts of cDNA solution per reaction with the following conditions: 95°C for 2 min, then 40 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min 40 s. Amplified RT-PCR fragments using H. piloselloides primers were cloned into the pGEM-T Easy vector (Promega) and sequenced to verify recovery of the expected product, and more than 97% identity to the original cDNA sequence was considered a match.
In Situ Hybridization Analysis of Selected Genes in Ovary Sections
In situ hybridization was carried out as previously described by Okada et al. (2007). Briefly, florets of H. praealtum R35 and H. pilosella P36 were collected and fixed in a buffered 4% (w/v) paraformaldehyde solution containing 0.25% glutaraldehyde and 1 mm DTT and embedded in BMM. Sections (3–4 µm) were hybridized with the digoxigenin-labeled sense and antisense RNA probes to detect transcripts from 9.45 RD22, 24.04 NLR, or 27.18 LOX genes (Supplemental Table S2; Roche Diagnostics). Hybridization signals were visualized by overnight incubation. Microscopy and photography were conducted as described by Okada et al. (2007).
454 Pyrosequencing of aRNA from Laser-Captured Samples
First-strand cDNA was synthesized using 20 µg of aRNA for each cell type with random hexamers, followed by second-strand cDNA synthesis using a BG-GsuI(dT) primer: 5′-GAGAGAGAGAATCCTTGCGGCCGCCTGGAG(T)16VN-3′ (V = A, G, or C). The polyA tail sequences were removed by GsuI digestion (Shibata et al., 2001), followed by phenol chloroform purification and size fractionation of cDNAs using a Sepharose G50 column to collect cDNA fragments greater than 150 bp. Due to the preliminary RNA amplification step, cDNA samples were enriched for short (<500 bp) fragments. 454 sequence data generation was conducted by the Australian Genome Research Facility, and the resulting sequence data sets have been deposited in the NCBI Sequence Read Archive public database (http://www.ncbi.nlm.nih.gov/sra; with BioSample accession nos. of SAMN01915662, SAMN01915663, and SAMN01915664).
Quality 454 Read Filtering, Pfam Search, and Enrichment Analysis
Raw 454 sequences were trimmed for adapter sequences, low quality, and ambiguous bases using the Lucy algorithm version 1.2.0 (Chou and Holmes, 2001) with default parameters. Trimmed sequences less than 100 bases were discarded. In total, 79.9% of raw sequences passed through quality filtering for use in assembly. All quality-filtered sequence reads were used to query the protein family database Pfam (http://pfam.sanger.ac.uk) to explore protein domain annotations predicted in the transcriptome sequence. The use of the larger set of sequence reads before assembly provided more direct access to individual read count information and also provided a means for analysis independent of the assembly algorithm. Sequence reads were translated in six frames, and putative peptides longer than 20 amino acids were analyzed against Pfam using a BLASTx E value threshold of 1E–5. Where multiple acceptable matches were found, each sequence was annotated by the best-scoring Pfam domain. Pfam domains observed in at least five reads were annotated for GO functional classes through the Pfam database and analyzed for enrichment in pairwise comparisons between the three cell types using the Fisher exact test (P value ≤ 0.05) with false discovery rate correction for multiple testing (Benjamini and Hochberg, 1995).
Contig Assembly and Gene Annotation
The Mimicking Intelligent Read Assembly algorithm version 3.2.0 (Chevreux et al., 2004) was used for assembly with the “accurate” parameter setting. The assembly resulted in 8,044 contigs for SO cells, 8,780 for AI cells, and 5,002 for EAE sacs. To generate the combined assembly, quality-filtered sequences from the three samples were combined and assembled, resulting in 18,219 contigs. These were further filtered to those that had an average read coverage across their length of at least three reads. Contig sequences from the combined assembly were compared against each cell type assembly, and sequences were considered matched if they had a BLASTn hit (E value ≤ 1E–10) across at least 80% of the smaller contig length. In cases where a combined assembly contig strongly matched multiple contigs from the same cell type, the match was only counted once. In this way, the combined assembly was used as a nonredundant reference to generate a representation of sequence similarity between cell types (Fig. 2A). The contigs were annotated by reciprocal BLAST to the NCBI nonredundant protein database and the TAIR10 peptide database. BLAST results were filtered to those with an E value of no more than 1E–5 and a match extending at least 80% of the query- or target-matched sequence. As Hieracium is a member of the Compositae family, the EST resource of the Compositae Genome Project sequence database (http://compgenomics.ucdavis.edu/) was queried for gene annotations. The NCBI Asteraceae (taxon identification: 4210) was also queried for annotations; however, the Compositae Genome Project database generated substantially more alignments and was used thereafter. Best-hit Compositae EST sequences were identified as those where Hieracium spp. contigs showed strong matches (BLASTn, E value ≤ 1E–10) across at least 80% of query or the Compositae EST with no more than one gap, which was shorter than 5% of total contig length. Best-hit Compositae EST sequences were annotated by comparison with the Arabidopsis TAIR10 peptide database by BLASTx. Enrichment of GO terms in pairwise comparisons between the three cell types was assessed by Fisher exact test (P value ≤ 0.05) with false discovery rate (Benjamini and Hochberg, 1995) adjustment using AgriGO (Du et al., 2010). A second-stage GO enrichment analysis was performed using the nEASE algorithm (Chittenden et al., 2012) and default parameters. nEASE aims to identify robust GO enrichments of more specific GO terms by borrowing strength across parent-child relationships in the GO ontology tree. An Expression Analysis Systematic Explorer (EASE) Arabidopsis ontology reference data set was created for this purpose from the GO annotations available at TAIR10.
Normalization of Sequence Reads to Number of the Contigs and Validation by Q-PCR
To compare abundance of contig sequences between cell types and for Q-PCR validation, the number of sequence reads attributable to each contig was normalized to reads per million per kilobase. An additional normalization factor was applied based on five ubiquitously expressed control genes, Elongation factor1-α, Polyubiquitin, Actin2, Ubiquitin-conjugating enzyme9, and Glyceraldehyde-3-P dehydrogenase C2, that are known to be stably expressed in different cells and tissues (Czechowski et al., 2005). Fifteen contigs and their normalized abundances were chosen for validation by Q-PCR. The first-strand cDNA synthesized from aRNA as described above was mixed with Absolute QPCR SYBR Green Mix (Thermo Fisher Scientific) and appropriate gene primers. Q-PCR was conducted in a Roter Gene RG-3000 (Corbett Life Science). All samples were measured in triplicate. The average expression of target genes was normalized as described by Rodrigues et al. (2008). These data are shown in Supplemental Figure S5.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Protocol S1. LCM, RNA isolation, and amplification.
Supplemental Figure S1. Validation of quality and quantity of aRNA from laser capture microdissected cell types from H. praealtum ovule sections.
Supplemental Figure S2. Validation of RT-PCR data obtained for low ovary-expressed genes in LCM samples by quantitative real-time PCR.
Supplemental Figure S3. Examination of two AI cell-detected genes, an abscisic acid-responsive RD22-like gene and a CC-NBS-LRR-like disease resistance gene, by in situ hybridization in ovules of sexual and apomictic plants.
Supplemental Figure S4. Flow chart of bioinformatic analyses of RNA sequencing data from each laser-captured ovule cell type with thresholds and filtering as shown.
Supplemental Figure S5. Comparison of the number of sequence reads in 15 genes assembled de novo and their expression in the three LCM samples by quantitative real-time PCR.
Supplemental Figure S6. Radar plots showing the similarity between Arabidopsis and apomictic H. praealtum ovule LCM data sets based on detection of TAIR10 transcripts on Agilent 4x44K arrays.
Supplemental Figure S7. Differentially expressed Arabidopsis homologs of H. praealtum contigs from AI cell, SO cell, and EAE sac data sets.
Supplemental Table S1. Isolation of RNA from laser-captured H. praealtum R35 apomictic ovule cell types and RNA yields after amplification.
Supplemental Table S2. Expression of low-level H. piloselloides (D18) ovary genes in amplified AI cell, SO cell, and EAE sac H. praealtum aRNA samples by RT-PCR.
Supplemental Table S3. Pfam enrichment analysis by reciprocal and pairwise comparisons between H. praealtum ovule cell types.
Supplemental Table S4. GO enrichment analysis by reciprocal and pairwise comparisons between H. praealtum ovule cell types.
Supplemental Table S5. Arabidopsis meiosis- and megaspore-associated genes examined for presence in H. praealtum AI cell contigs.
Supplemental Table S6. Discriminatory gene annotations associated with significant GO terms found through nested GO enrichment analysis between H. praealtum ovule cell types and comparison with Arabidopsis ovule genes on Agilent 4x44K arrays.
Acknowledgments
We thank the Australian Genome Research Facility for advice during transcript sequencing and Gwenda Mayo (Australian Centre for Plant Functional Genomics) for assistance with the Leica AS microscope.
Glossary
- AI
aposporous initial
- MMC
megaspore mother cell
- FM
functional megaspore
- EAE
early aposporous embryo
- LCM
laser capture microdissection
- SO
sporophytic ovule
- aRNA
amplified RNA
- cDNA
complementary DNA
- TAIR10
The Arabidopsis Information Resource 10
- NCBI
National Center for Biotechnology Information
- GO
Gene Ontology
- Q
quantitative
- SEA
singular Gene Ontology term enrichment analysis
- nEASE
nested Expression Analysis Systematic Explorer
- FG2-4
female gametophyte at megagametogenesis containing two to four nuclei
- DTT
dithiothreitol
- BMM
butyl methyl methacrylate
- RT
reverse transcription
References
- Acosta-García G, Vielle-Calzada JP. (2004) A classical arabinogalactan protein is essential for the initiation of female gametogenesis in Arabidopsis. Plant Cell 16: 2614–2628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertini E, Marconi G, Barcaccia G, Raggi L, Falcinelli M. (2004) Isolation of candidate genes for apomixis in Poa pratensis L. Plant Mol Biol 56: 879–894 [DOI] [PubMed] [Google Scholar]
- Armenta-Medina A, Demesa-Arévalo E, Vielle-Calzada JP. (2011) Epigenetic control of cell specification during female gametogenesis. Sex Plant Reprod 24: 137–147 [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. (2000) Gene ontology: tool for the unification of biology. Nat Genet 25: 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin MJ, Muskett P, Kahn K, Feys BJ, Jones JD, Parker JE. (2002) Regulatory role of SGT1 in early R gene-mediated plant defenses. Science 295: 2077–2080 [DOI] [PubMed] [Google Scholar]
- Baskin TI, Busby CH, Fowke LC, Sammut M, Gubler F. (1992) Improvements in immunostaining samples embedded in methacrylate - Localization of microtubules and other antigens throughout developing organs in plants of diverse taxa. Planta 187: 405–413 [DOI] [PubMed] [Google Scholar]
- Bencivenga S, Colombo L, Masiero S. (2011) Cross talk between the sporophyte and the megagametophyte during ovule development. Sex Plant Reprod 24: 113–121 [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc, B 57: 289–300 [Google Scholar]
- Carman JG. (1997) Asynchronous expression of duplicate genes in angiosperms may cause apomixis, bispory, tetraspory, and polyembryony. Biol J Linn Soc Lond 61: 51–94 [Google Scholar]
- Catanach AS, Erasmuson SK, Podivinsky E, Jordan BR, Bicknell R. (2006) Deletion mapping of genetic regions associated with apomixis in Hieracium. Proc Natl Acad Sci USA 103: 18650–18655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervigni GD, Paniego N, Pessino S, Selva JP, Díaz M, Spangenberg G, Echenique V. (2008) Gene expression in diplosporous and sexual Eragrostis curvula genotypes with differing ploidy levels. Plant Mol Biol 67: 11–23 [DOI] [PubMed] [Google Scholar]
- Chevreux B, Pfisterer T, Drescher B, Driesel AJ, Müller WE, Wetter T, Suhai S. (2004) Using the miraEST assembler for reliable and automated mRNA transcript assembly and SNP detection in sequenced ESTs. Genome Res 14: 1147–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chittenden TW, Howe EA, Taylor JM, Mar JC, Aryee MJ, Gómez H, Sultana R, Braisted J, Nair SJ, Quackenbush J, et al. (2012) nEASE: a method for gene ontology subclassification of high-throughput gene expression data. Bioinformatics 28: 726–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou HH, Holmes MH. (2001) DNA sequence quality trimming and vector removal. Bioinformatics 17: 1093–1104 [DOI] [PubMed] [Google Scholar]
- Couteau F, Belzile F, Horlow C, Grandjean O, Vezon D, Doutriaux MP. (1999) Random chromosome segregation without meiotic arrest in both male and female meiocytes of a dmc1 mutant of Arabidopsis. Plant Cell 11: 1623–1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR. (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day RC, Grossniklaus U, Macknight RC. (2005) Be more specific! Laser-assisted microdissection of plant cells. Trends Plant Sci 10: 397–406 [DOI] [PubMed] [Google Scholar]
- d’Erfurth I, Jolivet S, Froger N, Catrice O, Novatchkova M, Mercier R. (2009) Turning meiosis into mitosis. PLoS Biol 7: e1000124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- d’Erfurth I, Cromer L, Jolivet S, Girard C, Horlow C, Sun Y, To JP, Berchowitz LE, Copenhaver GP, Mercier R. (2010) The cyclin-A CYCA1;2/TAM is required for the meiosis I to meiosis II transition and cooperates with OSD1 for the prophase to first meiotic division transition. PLoS Genet 6: e1000989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drews GN, Koltunow AM. (2011) The female gametophyte. The Arabidopsis Book 9: e0155, /10.1199/tab.0155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z, Zhou X, Ling Y, Zhang Z, Su Z. (2010) agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res 38: W64-W70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberwine J, Yeh H, Miyashiro K, Cao Y, Nair S, Finnell R, Zettel M, Coleman P. (1992) Analysis of gene expression in single live neurons. Proc Natl Acad Sci USA 89: 3010–3014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Aguilar M, Michaud C, Leblanc O, Grimanelli D. (2010) Inactivation of a DNA methylation pathway in maize reproductive organs results in apomixis-like phenotypes. Plant Cell 22: 3249–3267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann NR. (2010) Apomixis and gene expression in Boechera. Plant Cell 22: 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honys D, Twell D. (2004) Transcriptome analysis of haploid male gametophyte development in Arabidopsis. Genome Biol 5: R85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juranič M, Srilunchang KO, Krohn NG, Leljak-Levanic D, Sprunck S, Dresselhaus T. (2012) Germline-specific MATH-BTB substrate adaptor MAB1 regulates spindle length and nuclei identity in maize. Plant Cell 24: 4974–4991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenneth NS, Duckett CS. (2012) IAP proteins: regulators of cell migration and development. Curr Opin Cell Biol 24: 871–875 [DOI] [PubMed] [Google Scholar]
- Kerk NM, Ceserani T, Tausta SL, Sussex IM, Nelson TM. (2003) Laser capture microdissection of cells from plant tissues. Plant Physiol 132: 27–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koltunow AM, Grossniklaus U. (2003) Apomixis: a developmental perspective. Annu Rev Plant Biol 54: 547–574 [DOI] [PubMed] [Google Scholar]
- Koltunow AM, Johnson SD, Bicknell RA. (1998) Sexual and apomictic development in Hieracium. Sex Plant Reprod 11: 213–230 [Google Scholar]
- Koltunow AM, Johnson SD, Bicknell RA. (2000) Apomixis is not developmentally conserved in related, genetically characterized Hieracium plants of varying ploidy. Sex Plant Reprod 12: 253–266 [Google Scholar]
- Koltunow AM, Johnson SD, Lynch M, Yoshihara T, Costantino P. (2001) Expression of rolB in apomictic Hieracium piloselloides Vill. causes ectopic meristems in planta and changes in ovule formation, where apomixis initiates at higher frequency. Planta 214: 196–205 [DOI] [PubMed] [Google Scholar]
- Koltunow AM, Johnson SD, Okada T. (2011b) Apomixis in hawkweed: Mendel’s experimental nemesis. J Exp Bot 62: 1699–1707 [DOI] [PubMed] [Google Scholar]
- Koltunow AMG, Johnson SD, Rodrigues JCM, Okada T, Hu Y, Tsuchiya T, Wilson S, Fletcher P, Ito K, Suzuki G, et al. (2011a) Sexual reproduction is the default mode in apomictic Hieracium subgenus Pilosella, in which two dominant loci function to enable apomixis. Plant J 66: 890–902 [DOI] [PubMed] [Google Scholar]
- Laspina NV, Vega T, Seijo JG, González AM, Martelotto LG, Stein J, Podio M, Ortiz JP, Echenique VC, Quarin CL, et al. (2008) Gene expression analysis at the onset of aposporous apomixis in Paspalum notatum. Plant Mol Biol 67: 615–628 [DOI] [PubMed] [Google Scholar]
- Li Y, Li T, Liu S, Qiu M, Han Z, Jiang Z, Li R, Ying K, Xie Y, Mao Y. (2004) Systematic comparison of the fidelity of aRNA, mRNA and T-RNA on gene expression profiling using cDNA microarray. J Biotechnol 107: 19–28 [DOI] [PubMed] [Google Scholar]
- Liu J, Zhang Y, Qin G, Tsuge T, Sakaguchi N, Luo G, Sun K, Shi D, Aki S, Zheng N, et al. (2008) Targeted degradation of the cyclin-dependent kinase inhibitor ICK4/KRP6 by RING-type E3 ligases is essential for mitotic cell cycle progression during Arabidopsis gametogenesis. Plant Cell 20: 1538–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonomura K, Miyoshi K, Eiguchi M, Suzuki T, Miyao A, Hirochika H, Kurata N. (2003) The MSP1 gene is necessary to restrict the number of cells entering into male and female sporogenesis and to initiate anther wall formation in rice. Plant Cell 15: 1728–1739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonomura K, Morohoshi A, Nakano M, Eiguchi M, Miyao A, Hirochika H, Kurata N. (2007) A germ cell specific gene of the ARGONAUTE family is essential for the progression of premeiotic mitosis and meiosis during sporogenesis in rice. Plant Cell 19: 2583–2594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien M, Major G, Chantha S, Matton DP. (2004) Isolation of S-RNase binding proteins from Solanum chacoense: identification of an SBP1 (RING finger protein) orthologue. Sex Plant Reprod 17: 81–87 [Google Scholar]
- Okada T, Catanach AS, Johnson SD, Bicknell RA, Koltunow AM. (2007) An Hieracium mutant, loss of apomeiosis 1 (loa1) is defective in the initiation of apomixis. Sex Plant Reprod 20: 199–211 [Google Scholar]
- Okada T, Ito K, Johnson SD, Oelkers K, Suzuki G, Houben A, Mukai Y, Koltunow AM. (2011) Chromosomes carrying meiotic avoidance loci in three apomictic eudicot Hieracium subgenus Pilosella species share structural features with two monocot apomicts. Plant Physiol 157: 1327–1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmedo-Monfil V, Durán-Figueroa N, Arteaga-Vázquez M, Demesa-Arévalo E, Autran D, Grimanelli D, Slotkin RK, Martienssen RA, Vielle-Calzada JP. (2010) Control of female gamete formation by a small RNA pathway in Arabidopsis. Nature 464: 628–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagnussat GC, Yu HJ, Ngo QA, Rajani S, Mayalagu S, Johnson CS, Capron A, Xie LF, Ye D, Sundaresan V. (2005) Genetic and molecular identification of genes required for female gametophyte development and function in Arabidopsis. Development 132: 603–614 [DOI] [PubMed] [Google Scholar]
- Pagnussat GC, Yu HJ, Sundaresan V. (2007) Cell-fate switch of synergid to egg cell in Arabidopsis eostre mutant embryo sacs arises from misexpression of the BEL1-like homeodomain gene BLH1. Plant Cell 19: 3578–3592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J, et al. (2012) The Pfam protein families database. Nucleic Acids Res 40: D290–D301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues JC, Cabral GB, Dusi DM, de Mello LV, Rigden DJ, Carneiro VT. (2003) Identification of differentially expressed cDNA sequences in ovaries of sexual and apomictic plants of Brachiaria brizantha. Plant Mol Biol 53: 745–757 [DOI] [PubMed] [Google Scholar]
- Rodrigues JC, Tucker MR, Johnson SD, Hrmova M, Koltunow AM. (2008) Sexual and apomictic seed formation in Hieracium requires the plant polycomb-group gene FERTILIZATION INDEPENDENT ENDOSPERM. Plant Cell 20: 2372–2386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Leal D, Vielle-Calzada JP. (2012) Regulation of apomixis: learning from sexual experience. Curr Opin Plant Biol 15: 549–555 [DOI] [PubMed] [Google Scholar]
- Schallau A, Arzenton F, Johnston AJ, Hähnel U, Koszegi D, Blattner FR, Altschmied L, Haberer G, Barcaccia G, Bäumlein H. (2010) Identification and genetic analysis of the APOSPORY locus in Hypericum perforatum L. Plant J 62: 773–784 [DOI] [PubMed] [Google Scholar]
- Schwarzacher T. (2003) Meiosis, recombination and chromosomes: a review of gene isolation and fluorescent in situ hybridization data in plants. J Exp Bot 54: 11–23 [DOI] [PubMed] [Google Scholar]
- Sharbel TF, Voigt ML, Corral JM, Galla G, Kumlehn J, Klukas C, Schreiber F, Vogel H, Rotter B. (2010) Apomictic and sexual ovules of Boechera display heterochronic global gene expression patterns. Plant Cell 22: 655–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharbel TF, Voigt ML, Corral JM, Thiel T, Varshney A, Kumlehn J, Vogel H, Rotter B. (2009) Molecular signatures of apomictic and sexual ovules in the Boechera holboellii complex. Plant J 58: 870–882 [DOI] [PubMed] [Google Scholar]
- Shi DQ, Liu J, Xiang YH, Ye D, Sundaresan V, Yang WC. (2005) SLOW WALKER1, essential for gametogenesis in Arabidopsis, encodes a WD40 protein involved in 18S ribosomal RNA biogenesis. Plant Cell 17: 2340–2354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi DQ, Yang WC. (2011) Ovule development in Arabidopsis: progress and challenge. Curr Opin Plant Biol 14: 74–80 [DOI] [PubMed] [Google Scholar]
- Shibata Y, Carninci P, Sato K, Hayatsu N, Shiraki T, Ishii Y, Arakawa T, Hara A, Ohsato N, Izawa M, et al. (2001) Removal of polyA tails from full-length cDNA libraries for high-efficiency sequencing. Biotechniques 31: 1042–1049 [DOI] [PubMed] [Google Scholar]
- Singh M, Burson BL, Finlayson SA. (2007) Isolation of candidate genes for apomictic development in buffelgrass (Pennisetum ciliare). Plant Mol Biol 64: 673–682 [DOI] [PubMed] [Google Scholar]
- Singh M, Goel S, Meeley RB, Dantec C, Parrinello H, Michaud C, Leblanc O, Grimanelli D. (2011) Production of viable gametes without meiosis in maize deficient for an ARGONAUTE protein. Plant Cell 23: 443–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith TF, Gaitatzes C, Saxena K, Neer EJ. (1999) The WD repeat: a common architecture for diverse functions. Trends Biochem Sci 24: 181–185 [DOI] [PubMed] [Google Scholar]
- Tucker MR, Araujo AC, Paech NA, Hecht V, Schmidt ED, Rossell JB, De Vries SC, Koltunow AM. (2003) Sexual and apomictic reproduction in Hieracium subgenus Pilosella are closely interrelated developmental pathways. Plant Cell 15: 1524–1537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker MR, Okada T, Hu Y, Scholefield A, Taylor JM, Koltunow AM. (2012a) Somatic small RNA pathways promote the mitotic events of megagametogenesis during female reproductive development in Arabidopsis. Development 139: 1399–1404 [DOI] [PubMed] [Google Scholar]
- Tucker MR, Okada T, Johnson SD, Takaiwa F, Koltunow AM. (2012b) Sporophytic ovule tissues modulate the initiation and progression of apomixis in Hieracium. J Exp Bot 63: 3229–3241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker MR, Paech NA, Willemse MTM, Koltunow AMG. (2001) Dynamics of callose deposition and β-1,3-glucanase expression during reproductive events in sexual and apomictic Hieracium. Planta 212: 487–498 [DOI] [PubMed] [Google Scholar]
- Vielle-Calzada JP, Nuccio ML, Budiman MA, Thomas TL, Burson BL, Hussey MA, Wing RA. (1996) Comparative gene expression in sexual and apomictic ovaries of Pennisetum ciliare (L.) Link. Plant Mol Biol 32: 1085–1092 [DOI] [PubMed] [Google Scholar]
- Vierstra RD. (2012) The expanding universe of ubiquitin and ubiquitin-like modifiers. Plant Physiol 160: 2–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb M, Gunning B. (1994) Embryo sac development in Arabidopsis thaliana II. The cyto skeleton during megagametogenesis. Sex Plant Reprod 7: 153–163 [Google Scholar]
- Wuest SE, Vijverberg K, Schmidt A, Weiss M, Gheyselinck J, Lohr M, Wellmer F, Rahnenführer J, von Mering C, Grossniklaus U. (2010) Arabidopsis female gametophyte gene expression map reveals similarities between plant and animal gametes. Curr Biol 20: 506–512 [DOI] [PubMed] [Google Scholar]
- Xie DX, Feys BF, James S, Nieto-Rostro M, Turner JG. (1998) COI1: an Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280: 1091–1094 [DOI] [PubMed] [Google Scholar]
- Zhang S, Cao J, Kong YM, Scheuermann RH. (2010) GO-Bayes: Gene Ontology-based overrepresentation analysis using a Bayesian approach. Bioinformatics 26: 905–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, de Palma J, Oane R, Gamuyao R, Luo M, Chaudhury A, Hervé P, Xue Q, Bennett J. (2008) OsTDL1A binds to the LRR domain of rice receptor kinase MSP1, and is required to limit sporocyte numbers. Plant J 54: 375–387 [DOI] [PMC free article] [PubMed] [Google Scholar]