A microarray analysis reveals 828 unique transcripts with expression related to crown root initiation during coleoptilar node development.
Abstract
Maize (Zea mays) develops an extensive shoot-borne root system to secure water and nutrient uptake and to provide anchorage in the soil. In this study, early coleoptilar node (first shoot node) development was subjected to a detailed morphological and histological analysis. Subsequently, microarray profiling via hybridization of oligonucleotide microarrays representing transcripts of 31,355 unique maize genes at three early stages of coleoptilar node development was performed. These pairwise comparisons of wild-type versus mutant rootless concerning crown and seminal roots (rtcs) coleoptilar nodes that do not initiate shoot-borne roots revealed 828 unique transcripts that displayed RTCS-dependent expression. A stage-specific functional analysis revealed overrepresentation of “cell wall,” “stress,” and “development”-related transcripts among the differentially expressed genes. Differential expression of a subset of 15 of 828 genes identified by these microarray experiments was independently confirmed by quantitative real-time-polymerase chain reaction. In silico promoter analyses revealed that 100 differentially expressed genes contained at least one LATERAL ORGAN BOUNDARIES domain (LBD) motif within 1 kb upstream of the ATG start codon. Electrophoretic mobility shift assay experiments demonstrated RTCS binding for four of these promoter sequences, supporting the notion that differentially accumulated genes containing LBD motifs are likely direct downstream targets of RTCS.
The complex root system of maize (Zea mays) consists of embryonic primary and seminal roots and shoot-borne crown and brace roots that appear from consecutive belowground and aboveground nodal structures of the shoot (Hochholdinger et al., 2004a; Hochholdinger, 2009). All root types have the potential to form lateral roots from dividing pericycle cells postembryonically (Hochholdinger et al., 2004b). The shoot-borne root system, which comprises the major portion of the adult root stock, prevents lodging of maize plants and ensures adequate uptake of water and nutrients (Hochholdinger et al., 2004b). In total, adult maize plants develop approximately 70 shoot-borne roots, which are organized on average in six whorls of underground crown roots and two to three whorls of aboveground brace roots (Hochholdinger, 2009), although there is genetic variability for this trait. The first whorl of crown roots is initiated from the first shoot node, the coleoptilar node, approximately 4 to 6 d after germination, opposite to collateral vascular bundles (Hochholdinger, 2009).
The maize mutant rootless concerning crown and seminal roots (rtcs) was identified by its increased lodging in field trials (Hetz et al., 1996). Subsequent analyses revealed that rtcs mutants do not initiate seminal and shoot-borne roots (Hetz et al., 1996). Hence, the rtcs mutant forms only a primary root and lateral roots derived from the primary root. Nevertheless, rtcs plants can grow to maturity under controlled greenhouse conditions (Hetz et al., 1996).
The rtcs gene (GRMZM2G092542) encodes a member of the Lateral Organ Boundaries (LOB) domain (LBD) transcription factor family (Taramino et al., 2007) and is localized in the nucleus (Majer et al., 2012). The conserved LBD is plant specific and contains a C block that is predicted to contain a secondary structure that resembles a DNA-binding zinc finger (Majer and Hochholdinger, 2011). It has been demonstrated that LBD proteins recognize a specific 5′-GCGGCG-3′ DNA consensus sequence termed the LBD motif (Husbands et al., 2007). The RTCS protein can bind to an LBD motif of the arf34 gene, while the ARF34 protein can bind to an LBD motif in the rtcs promoter (Majer et al., 2012). Moreover, LBD proteins contain a C-terminal GAS block that forms a coiled-coil structure, which is predicted to be a Leu zipper motif required for protein-protein interactions (Majer and Hochholdinger, 2011). It has also been demonstrated by yeast two-hybrid experiments that the RTCS protein can homointeract with RTCS and heterointeract with its closely related homolog RTCS-like (Majer et al., 2012).
LBD proteins are involved in various developmental processes. Mutations in LBD genes affect leaf venation in Arabidopsis (Arabidopsis thaliana; Iwakawa et al., 2002), patterning of stem cells in axillary meristems of maize (Bortiri et al., 2006), shoot-borne root formation in rice (Oryza sativa) and maize (Inukai et al., 2005; Liu et al., 2005; Taramino et al., 2007), and glume formation in rice (Li et al., 2008). Recently, it has been demonstrated in Arabidopsis that LBD16/ASYMMETRIC LEAVES18 (ASL18) is involved in the regulation of lateral root formation downstream of ARF7 and ARF19 (Lee et al., 2009).
The objective of this study was to identify differentially expressed genes between wild-type and rtcs mutant coleoptilar nodes at different developmental stages that are likely to be regulated by RTCS, with the goal of better understanding the molecular framework associated with crown root formation in maize.
RESULTS
Morphological and Histological Characterization of Coleoptilar Node Development in the Wild Type and the Crown Root-Deficient Mutant rtcs
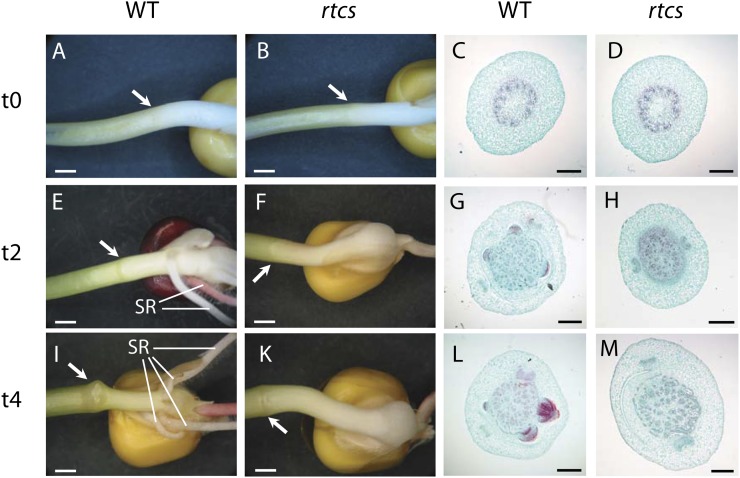
Wild-type seedlings initiate shoot-borne roots at the coleoptilar node 5 to 10 d after germination (Sauer et al., 2006). In contrast, rtcs mutant seedlings completely fail to initiate shoot-borne roots (Hetz et al., 1996). To investigate the early stages of shoot-borne root initiation in more detail, we defined the stages of coleoptilar node development based on the first appearance of this structure in seedlings. In both rtcs and wild-type seedlings, the coleoptilar node is formed 4 to 6 d after germination. To account for this variability, the time of coleoptilar node formation was defined as t0 for subsequent analyses. At this developmental stage, the gross morphology and transverse sections of the coleoptilar nodes of wild-type and rtcs seedlings cannot be distinguished (Fig. 1, A–D). For subsequent molecular analyses, we selected coleoptilar nodes 2 d (t2) and 4 d (t4) after coleoptilar node appearance. At t2, wild-type and rtcs coleoptilar nodes are still morphologically indistinguishable (Fig. 1, E and F). However, cross sections demonstrated that at this stage, histological differences were manifested by shoot-borne root primordia that had been initiated in wild-type seedlings (Fig. 1G) but that were absent in rtcs seedlings (Fig. 1H). At t4, rtcs coleoptilar nodes (Fig. 1K) lack the bulges in the vicinity of the coleoptilar node (Fig. 1I) that indicate the imminent appearance of crown roots in wild-type seedlings. In line with this observation, cross sections of t4 coleoptilar nodes of the wild type display emerging crown roots (Fig. 1L), while these structures are absent in the rtcs mutant seedlings (Fig. 1M). Hence, histological analyses using the time point of coleoptilar node appearance as a reference allowed us to precisely define distinct developmental stages of the development of crown root primordia.
Figure 1.
Morphological (A, B, E, F, I, and K) and histological (C, D, G, H, L, and M) features of maize wild-type (WT: A, C, E, G, I, and L) and rtcs (B, D, F, H, K, and M) coleoptilar nodes during node appearance (t0; A–D), 2 d after node appearance (t2; E–H), and 4 d after node appearance (t4; I–M). Coleoptilar nodes are indicated by white arrows. SR, Seminal roots. For details, see text. Bars = 2.5 mm (A, B, E, F, I, and K) and 500 µm (C, D, G, H, L, and M). [See online article for color version of this figure.]
Microarray Profiling of RTCS-Dependent Gene Expression during Crown Root Formation in the Coleoptilar Node
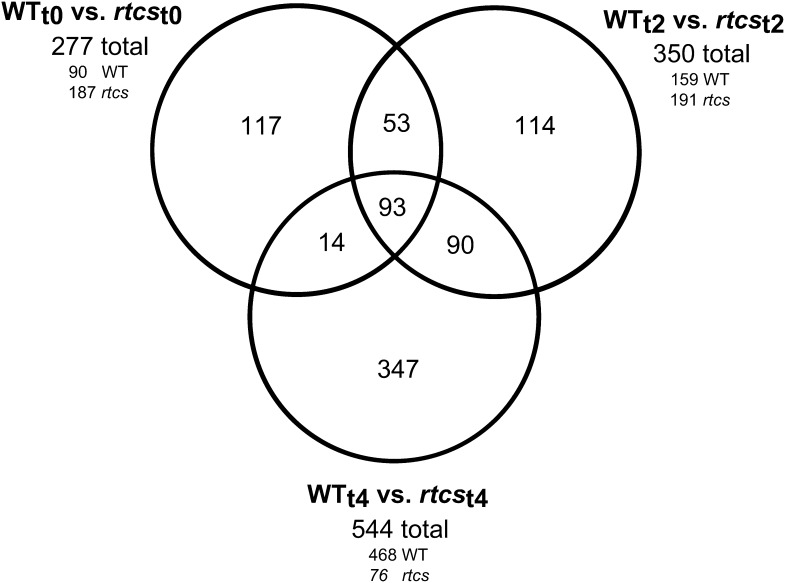
To identify genes regulated by RTCS during crown root formation in the coleoptilar node, microarray experiments were performed using a maize microarray platform containing 65,646 different 60-mer oligonucleotide features that represent transcripts of 31,335 unique maize genes (see “Materials and Methods”; Supplemental Table S1). These experiments compared gene expression between wild-type and rtcs seedlings during three stages of coleoptilar node development (WTt0 versus rtcst0, WTt2 versus rtcst2 and WTt4 versus rtcst4) based on the histological experiments described in Figure 1. In these pairwise comparisons, 958 (of 65,646) microarray features were differentially expressed (fold change ≥ 2, false discovery rate ≤ 5%) at least during one developmental stage (Supplemental Table S2). The 958 differential microarray features represented 828 transcripts covered by unique genes, with 727 transcripts covered by one microarray feature and 101 transcripts represented by two or more microarray features. Among the differentially expressed genes, the expression trend (i.e. preferential expression in the wild type or rtcs) was perfectly conserved between different microarray features corresponding to a unique transcript (Supplemental Table S2). In total, 93 of the 828 unique transcripts were differentially expressed at all three developmental stages between wild-type and mutant rtcs (Fig. 2). Hence, while most differentially expressed genes were regulated by RTCS in a development-dependent manner, 93 unique transcripts were regulated by RTCS constitutively at all three stages of early coleoptilar node development analyzed in this study. The highest number of stage-specific expression differences (544 unique transcripts) was observed at t4 (Fig. 2), while only 277 differentially accumulated unique transcripts were observed at t0 and 350 at t2. Hence, the number of differentially accumulated transcripts coincided with the development of shoot-borne root primordia in wild-type coleoptilar nodes between t0 and t4 that are absent in rtcs (Fig. 1).
Figure 2.
Differential expression of 828 unique genes between wild-type and rtcs coleoptilar nodes at three developmental stages (t0, t2, and t4). The Venn diagram illustrates the number of differentially expressed genes for all three pairwise comparisons and the number of genes overlapping between the comparisons. For each developmental stage, the total number of differentially expressed genes and the number of genes preferentially expressed in the wild type (WT) and rtcs are indicated.
The 828 differentially expressed genes between the wild type and rtcs (Fig. 2) were functionally annotated using the automated annotation pipeline Mercator (Supplemental Table S2). In total, 149, 194, and 307 unique differentially expressed transcripts were annotated at stages t0, t2, and t4, respectively. Subsequently, functional categories that are significantly overrepresented among the differentially expressed genes were identified (Table I). At all three stages, the category “miscellaneous” was significantly overrepresented. This category contains all genes that were not more specifically classified. These genes will not be further discussed. At t0 (i.e. before the initiation of shoot-borne roots in wild-type coleoptilar nodes), no other functional category was overrepresented among the differentially expressed genes. In contrast, at stages t2 and t4, genes related to “cell wall synthesis” were overrepresented among the differentially expressed genes; these genes may be associated with the formation of the crown root primordia during these stages. Finally, at stage t2, transcripts related to “stress” were overrepresented among the differentially expressed genes, while at stage t4 (i.e. shortly before the appearance of crown roots), transcripts related to “development” were overrepresented.
Table I. Assignment of differentially expressed genes to the main functional categories (MapMan).
Functional categories containing more or fewer genes than expected (based on the distribution of functional categories of all genes represented on the microarray chip) are highlighted by + or −.
| Bin Code | Bin Namea | All Genesb | Wild Type versus rtcs t0 | Directionc | Pd | Wild Type versus rtcs t2 | Direction | P | Wild Type versus rtcs t4 | Direction | P |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Photosynthesis | 274 | 1 | 1 | |||||||
| 2 | Major CHO metabolism | 169 | 1 | 3 | 1 | ||||||
| 3 | Minor CHO metabolism | 171 | 1 | 1 | 4 | ||||||
| 4 | Glycolysis | 77 | |||||||||
| 5 | Fermentation | 30 | 1 | 2 | 2 | ||||||
| 6 | Gluconeogenesis/glyoxylate cycle | 17 | |||||||||
| 7 | OPP | 48 | |||||||||
| 8 | TCA/organic transformation | 125 | 1 | 1 | |||||||
| 9 | Mitochondrial electron-transport/ATP synthesis | 157 | 1 | 1 | |||||||
| 10 | Cell wall | 552 | 7 | 12 | + | <0.01 | 19 | + | <0.01 | ||
| 11 | Lipid metabolism | 560 | 4 | 3 | 6 | ||||||
| 12 | Nitrogen metabolism | 50 | 1 | ||||||||
| 13 | Amino acid metabolism | 383 | 2 | 1 | 2 | ||||||
| 14 | Sulfur assimilation | 15 | |||||||||
| 15 | Metal handling | 73 | 1 | 2 | |||||||
| 16 | Secondary metabolism | 462 | 3 | 4 | 6 | ||||||
| 17 | Hormone metabolism | 586 | 5 | 5 | 12 | 0.23 | |||||
| 18 | Cofactor and vitamin metabolism | 88 | |||||||||
| 19 | Tetrapyrrole synthesis | 80 | 1 | ||||||||
| 20 | Stress | 866 | 9 | 14 | + | 0.03 | 18 | 0.12 | |||
| 21 | Redox regulation | 251 | 1 | ||||||||
| 22 | Polyamine metabolism | 24 | 1 | ||||||||
| 23 | Nucleotide metabolism | 224 | 1 | ||||||||
| 24 | Biodegradation of xenobiotics | 15 | 2 | 2 | |||||||
| 25 | C1 metabolism | 35 | 1 | 1 | |||||||
| 26 | Miscellaneous | 1,705 | 24 | + | <0.01 | 33 | + | <0.01 | 66 | + | <0.01 |
| 27 | RNA | 4,117 | 27 | 0.48 | 32 | 0.24 | 64 | 0.58 | |||
| 28 | DNA | 777 | 6 | 4 | 9 | ||||||
| 29 | Protein | 4,699 | 28 | 0.20 | 31 | − | 0.03 | 30 | − | <0.01 | |
| 30 | Signaling | 1,542 | 7 | 11 | 0.34 | 14 | 0.07 | ||||
| 31 | Cell | 1,025 | 7 | 8 | |||||||
| 32 | MicroRNA, natural antisense | 1 | 7 | ||||||||
| 33 | Development | 657 | 5 | 11 | 0.05 | 19 | + | <0.01 | |||
| 34 | Transport | 1,340 | 10 | 0.99 | 17 | 0.21 | 19 | 0.90 | |||
| 35 | Not assigned | 16,256 | 128 | 0.40 | 156 | 0.76 | 237 | 0.99 | |||
| Sume | 37,451 | 279 | 353 | 546 |
Only the 35 main functional categories were considered. bFunctional classification of all genes present on the microarray chip. cFor direction, the column indicates if the functional category is overrepresented (+) or underrepresented (−) based on the comparison of detected and expected numbers of differentially expressed genes. The expected number is calculated based on the functional distribution of all genes on the microarray chip. dP values are based on a χ2 test for independence with Yates’ continuity correction. eAll genes that were assigned to a functional category by the MapMan software are summed up. This number is larger than the number of genes in the considered subset because numerous genes are assigned to more than one functional category. CHO, Carbohydrate; OPP, oxidative pentosephosphate pathway.
Confirmation of Differential Expression by Quantitative Real-Time PCR
To independently confirm the microarray hybridization results, a subset of 15 transcripts that corresponded to differentially expressed microarray features was subjected to quantitative real-time (qRT)-PCR analyses (Supplemental Table S3). Among these, 11 genes were preferentially expressed in wild-type coleoptilar nodes at all three developmental stages (t0, t2, and t4), while two genes were preferentially expressed in wild-type coleoptilar nodes at two developmental stages (GRMZM2G053493_T01 at t2 and t4 and GRMZM2G165972_T02 at t0 and t4) and another two genes at one developmental stage (GRMZM2G070603_T01 and GRMZM2G150374_T04 at t4). For all 39 gene/stage combinations tested for the 15 genes, preferential expression in wild-type coleoptilar nodes was confirmed by qRT-PCR, thus supporting the microarray results. For 31 of 39 gene/stage combinations, qRT-PCR experiments confirmed fold change ≥ 2 and statistical significance according to Student’s t test (Supplemental Table S3).
Identification of Putative RTCS Target Genes
Genes differentially expressed between wild-type and rtcs coleoptilar nodes are likely directly or indirectly regulated by RTCS. Previous studies suggested that LBD proteins can directly bind to the 6-bp consensus LBD motif 5′-GCGGCG-3′ (Husbands et al., 2007). Among 828 differentially expressed genes, 100 RTCS-regulated genes contained between one and 11 LBD motifs within 1 kb upstream of the ATG start codon (Supplemental Table S4). Among those, 65 genes were preferentially expressed in wild-type coleoptilar nodes while 35 where preferentially expresses in rtcs coleoptilar nodes.
The abundance of LBD motifs in the 1-kb region immediately upstream of the ATG start codon of each of 39,656 high-confidence maize gene models was determined. We then compared the distributions of LBD motifs in the differentially expressed genes with those of all expressed genes minus the differentially expressed maize genes (Supplemental Table S5). While 12% of the differentially expressed genes contain one or more LBD promoter element, 28% of all maize genes contain one or more LBD motifs in their promoter. The dichotomized frequency distribution of the number of LBD motifs (zero and one or more) per gene between differentially expressed genes and the remaining genes among all 39,656 genes was compared (see “Materials and Methods”). This analysis indicated that genes without an LBD promoter element are overrepresented among the differentially expressed genes (P ≤ 0.0001). These results could be explained by the observation that LBD genes are mainly regulatory genes expressed at low levels that might not allow identifying them as differentially expressed in microarray experiments. Moreover, this might imply an overrepresentation of differentially expressed genes that are not direct targets of RTCS but that act farther downstream of RTCS.
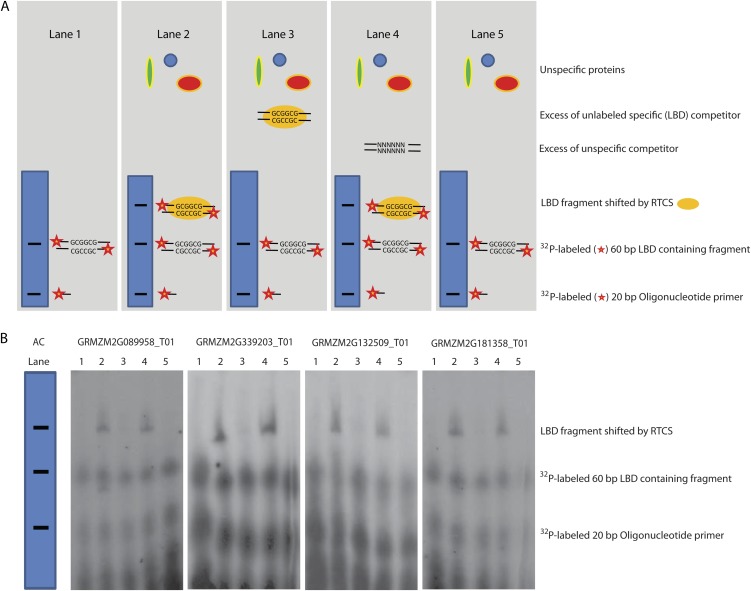
Direct RTCS interaction with LBD promoter sequences was assayed for four differentially expressed genes by electrophoretic mobility shift assay (EMSA) experiments. In all EMSA experiments, the LBD motif that was proximal to the ATG start codon was surveyed. The analyzed genes included a galactosyltransferase (GRMZM2G181358_T01) and a xylanase inhibitor, TAXI-IV (GRMZM2G132509_T01), each containing two LBD motifs, a stress response protein gene that contains eight LBD motifs (GRMZM2G089958_T01), and an ATP-binding protein gene that contains 11 LBD motifs (GRMZM2G339203_T01) and displays similarity to a gene encoding a casein kinase. For EMSA experiments, 55- to 60-bp promoter sequences containing the LBD motif in a central position were used (Supplemental Table S6). For all four genes, binding of RTCS to these promoter sequences was demonstrated (Fig. 3, lane 2). The specificity of RTCS binding was demonstrated by specific competition with a 50-fold excess of unlabeled LBD promoter sequences that outcompeted the labeled promoter sequences, thus reducing the shifted band (Fig. 3, lane 3). Unspecific competition by herring sperm DNA, which did not interact with RTCS, did not affect the shift (Fig. 3, lane 4). To confirm that RTCS rather than another protein of the bacterial protein lysate binds to the subjected DNA oligonucleotides, crude protein lysate from Escherichia coli BL21 cells expressing only the glutathione S-transferase (GST) tag instead of the GST-tagged RTCS protein was applied as a negative control (Fig. 3, lane 5). Finally, to exclude the possibility that multimerization of oligonucleotide fragments led to the observed shifts, fragments containing labeled LBD were loaded on the gel (Fig. 3, lane 1). In summary, these experiments demonstrated that RTCS can bind in vitro to the selected LBD promoter sequences. Subsequent genetic analyses of these candidate genes will be required to confirm these putative downstream targets of RTCS.
Figure 3.
EMSAs of radioactively labeled promoter fragments of four genes containing central 5′-GCGGCG-3′ LBD motifs that are bound by recombinant RTCS proteins. A, Schematic of the components loaded to each of the five lanes and the expected bands for the experiments depicted in B. Lane 1 contains 60-bp 32P-labeled 55- to 60-bp PCR-amplified promoter sequences and 20-bp 32P-labeled oligonucleotide primers that were not incorporated into 60-bp fragments during PCR. These two types of DNA fragments result in two bands on the gel. In lane 2, protein extract containing overexpressed recombinant RTCS and unspecific proteins are added. RTCS binds to the labeled LBD containing 60-bp PCR fragments, leading to a shift and a third band. Lane 3 represents a control experiment that demonstrates specific binding of RTCS to the LBD domain by introducing a 50× excess of nonlabeled, and thus invisible, 60-bp LBD-containing PCR product. Lane 4 illustrates another control. In this lane, a 50× excess of unspecific genomic herring sperm DNA was introduced. Since this DNA does not contain LBD motifs, RTCS still binds to radioactively labeled PCR products that contain LBD motifs. Lane 5 is the third control lane, which contains the same unspecific proteins as in lanes 2 to 4. However, in this lane, no overexpressed RTCS is included in the protein extract, demonstrating that the shift is not conditioned by unspecific protein binding. B, Specific binding of RTCS to four LBD-containing promoter sequences of genes that were differentially expressed in microarray and qRT-PCR experiments between wild-type and rtcs coleoptilar nodes (Supplemental Table S3). For all four genes, the expected band pattern for the five lanes is observed, suggesting in vitro binding of RTCS to these promoter sequences. AC, Accession. [See online article for color version of this figure.]
DISCUSSION
Histological Dissection of the Early Stages of Coleoptilar Node Development in the Wild Type and rtcs
Maize wild-type and mutant rtcs coleoptilar nodes can be distinguished by their capacity to initiate shoot-borne roots (Hetz et al., 1996). Wild-type seedlings initiate crown roots approximately 5 d after germination, and these roots appear approximately 10 d after germination (Sauer et al., 2006). The germination reference system introduces a lot of variability, because coleoptilar nodes appear between 4 and 6 d after germination (Sauer et al., 2006). Therefore, in this study, the appearance of the coleoptilar node was chosen as reference time point t0 for crown root development. This allows a more accurate determination of coleoptilar and crown root development and makes the results of the transcriptome analyses associated with crown root development more meaningful.
RTCS-Dependent Regulation of Gene Expression during Crown Root Development
Comparative transcriptome profiling of the three developmental stages of coleoptilar node development between the wild type and the mutant rtcs resulted in 828 unique genes that were differentially expressed between the two genotypes at least at one developmental stage. When examining differential gene expression, an increasing number from 277 (t0) to 350 (t2) to 544 (t4) differentially expressed genes were observed during the progress of coleoptilar node development. These numbers coincide with increasing morphological differences between wild-type and rtcs coleoptilar nodes during early development (Sauer et al., 2006). Similarly, the relative increase of differentially expressed genes preferentially expressed in wild-type coleoptilar nodes between t0 and t4 might be associated with the emergence of crown root primordia in wild-type coleoptilar nodes. At t2 and t4, a subset of differentially expressed genes might be specifically expressed in crown root primordia, which would result in differentially expressed genes preferentially expressed in wild-type coleoptilar nodes. Remarkably, already at t0 (i.e. prior to observable histological differences between the two genotypes), a considerable number of differentially expressed genes were detected. Likely, these genes are associated with the dedifferentiation of cortex cells (Hoppe et al., 1986), an RTCS-dependent process that precedes the first cell divisions in the coleoptilar node. Similar results were obtained in a comparative transcriptome survey of the wild type and the lateral root-deficient mutant rum1 (Woll et al., 2005). In that study, a significant number of differentially expressed genes were observed in pericycle cells prior to lateral root initiation, and it has been suggested that these genes may be associated with the early stages of lateral root initiation prior to the first cell divisions (Woll et al., 2005). Similarly, significant differences in global gene expression levels have also been observed in primary roots of different maize inbred lines and hybrids prior to the manifestation of morphological differences between the roots of these genotypes (Hoecker et al., 2008; Paschold et al., 2012). The increasing numbers of differentially expressed genes after the initial cell divisions in the crown root primordia (t2) and shortly before the appearance of the crown roots (t4) correspond to the increasing histological differences in the coleoptilar nodes of the wild type and rtcs that are a result of the cell divisions leading to crown roots in wild-type coleoptilar nodes (Hoppe et al., 1986). A set of 93 genes display RTCS-dependent expression throughout all three stages of early crown root formation in the coleoptilar node. Hence, these genes are controlled by RTCS throughout coleoptilar node development independent of the developmental status of the coleoptilar node, implying their importance for crown root formation at all stages of this developmental process.
Other than the “miscellaneous” category, no other functional category was overrepresented at t0, when no histological differences were apparent between the wild type and rtcs. The multifaceted character of the miscellaneous category impedes inferences toward specific cellular processes and implies that a variety of functions are involved in the specification of maize coleoptilar nodes for crown root initiation. At t2 and t4 (i.e. when crown root primordia are formed in wild-type nodes), differentially expressed genes associated with cell wall biosynthesis were overrepresented. This is in line with the observed phenotypes in the coleoptilar node. Cell divisions leading to crown root meristems provide a large number of new meristematic cells, a process that requires enhanced cell wall biosynthesis. In support of this notion, the mutant lrt1, which is defective in lateral root formation, displays cell wall-related defects, including lignification in peripheral layers, the deposition of polyphenolic substances, and a higher activity of peroxidases (Husakova et al., 2013). In addition, at t2, stress-related genes are overrepresented among the differentially expressed genes. At early stages of development, the embryonic primary and seminal roots provide water and nutrients for the growing seedling (Hochholdinger et al., 2004a). These embryonic roots are later supported by the emerging crown roots (Hochholdinger et al., 2004b). The mutant rtcs does not form seminal and crown roots (Hetz et al., 1996). Apparently, at stage t2, the supply of the plant by the primary root might not be sufficient, which could lead to the induction of stress-related genes and the overrepresentation of this category among the differentially expressed genes in the coleoptilar node at the shoot-root boundary. This is of particular relevance because it implies that shortly before crown root formation (t2), seminal roots could sufficiently supply the coleoptilar node in wild-type plants, a process that is impaired in rtcs due to the absence of seminal roots. In this context, it should be noted that the number of seminal roots is quite variable between different genotypes (Hochholdinger et al., 2004b). This implies that even if a genotype with few seminal roots would suffer some stress due to the lack of sufficient water and nutrient supply at t2, this could be compensated by the appearance of shoot-borne roots a few days later. This is of relevance for the overall condition of the plant, as the early stages of root development already predetermine the vigor of the adult root systems and, thus, plant development, as demonstrated for 40 different maize genotypes (Nass and Zuber, 1971).
Moreover, the functional category of “development” was overrepresented at t4. This is not surprising given the fact that wild-type and rtcs coleoptilar nodes differ in the developmental process of crown root formation, which are close to appearance at this stage. Hence, the genes differentially expressed in this category might be directly related to the development of crown roots.
RTCS-Dependent Regulation of LOB Domain Genes and Transcription Factors in the Coleoptilar Node
LOB genes constitute a plant-specific family of transcription factors comprising 43 members in Arabidopsis (Matsumura et al., 2009), 35 members in rice (Yang et al., 2006), and 43 members in maize (Majer and Hochholdinger, 2011) that control different aspects of plant development. Members of this gene family can be divided into different classes and subtypes, which are involved in distinct aspects of plant development (Majer and Hochholdinger, 2011). Most LBD genes studied thus far are involved in aboveground development, including adaxial-abaxial patterning of Arabidopsis leaves (Lin et al., 2003), proximal-distal patterning in Arabidopsis petals (Chalfun et al., 2005), embryo sac and leaf development in maize (Evans, 2007), leaf expansion in Arabidopsis (Iwakawa et al., 2007), and glume formation in rice (Li et al., 2008). However, it has also been demonstrated that LBD genes are involved in different aspects of root formation, including lateral root formation in Arabidopsis (Okushima et al., 2007), shoot-borne root formation in rice (Inukai et al., 2005; Liu et al., 2005), and shoot-borne and seminal root initiation in maize (Taramino et al., 2007). Mutations in the maize LBD gene rtcs result in plants that do not initiate shoot-borne and seminal roots (Hetz et al., 1996).
Although the interactions of LBD genes remain largely elusive to date, the differential expression of several LBD transcription factors between wild-type and rtcs coleoptilar nodes and during different stages of wild-type coleoptilar node development suggests a role of some of these genes in specification of the coleoptilar node and RTCS-dependent specification of shoot-borne root development. The maize LBD genes rtcs-neighbour (rtcn; GRMZM2G092483_T02; Taramino et al., 2007) and lbd34 (GRMZM2G075499_T01; Majer et al., 2012) are preferentially expressed in wild-type versus rtcs coleoptilar nodes at all three developmental stages dissected in this study. rtcn is physically located next to the rtcs gene in the maize genome (Taramino et al., 2007). Although rtcs and rtcn display complementary expression levels in most tissues, rtcn revealed an expression of approximately 140 ppm in a massively parallel signature sequencing analysis of the nodal root tip meristem (Taramino et al., 2007). This result is consistent with higher rtcn expression in meristem-forming wild-type versus meristemless rtcs coleoptilar nodes. A phylogenetic analysis of maize, rice, and Arabidopsis LBD genes revealed that the four maize LBD genes rtcs, rtcl, rtcn, and lbd34 belong to a subclade of the extensive gene family (Majer and Hochholdinger, 2011).
The rtcs gene (GRMZM2G092542) was represented by two microarray features, A4242726 and A4259513, which, however, were not among the 958 differential microarray features. Both microarray features displayed for the first two developmental stages, t0 and t2, preferential expression in wild-type roots, with similar fold change (2 or less), and at stage t4, preferential expression in the wild type, with fold change of 2.1 and 2.3. However, at all developmental stages, there was for both features not enough statistical power (false discovery rate > 5%) to declare them significantly different. In a subsequent qRT-PCR experiment, preferential expression of rtcs in WTt4 versus rtcst4 was demonstrated (Supplemental Table S3). Moreover, the maize LBD gene asl2 (Alexandrov et al., 2009; GRMZM2G154320_T01) was preferentially expressed in wild-type versus rtcs coleoptilar nodes at all three developmental stages. Although this gene has not yet been characterized, its constitutive control by RTCS at different stages of coleoptilar node development suggests that ASL2 could be involved in crown root development.
Another transcription factor that was almost exclusively expressed in wild-type coleoptilar nodes at all three analyzed stages and thus might have a role in RTCS-dependent development of the coleoptilar node or shoot-borne roots is baby boom1 (Alexandrov et al., 2009; GRMZM2G366434_T01), a transcription factor belonging to the APETALA2/ ETHYLENE-RESPONSIVE ELEMENT-BINDING PROTEIN (AP2/EREBP) family. Its closest homolog in Arabidopsis is PLETHORA2 (Aida et al., 2004), which, together with PLETHORA1, is required for the formation of root stem cells and roots (Aida et al., 2004), implying a similar RTCS-dependent role of these genes in shoot-borne root formation. Moreover, heat shock-complementing factor1 (Gardiner et al., 2004; NP_001106068), which shows high sequence homology to PLETHORA2, is also expressed preferentially in wild-type coleoptilar nodes at all three developmental stages assayed in this study. Another member of the AP2/EREBP family of transcription factors that is preferentially expressed in wild-type coleoptilar nodes during all three stages of development is branched silkless1 (bd1), which controls spikelet meristem formation in maize (Chuck et al., 2002). The rtcs gene is also expressed in spikelets (Taramino et al., 2007) and in rice spikelets has been demonstrated to have the competence to form roots under stress conditions (Jones and Pope, 1942; Taniguchi and Futsuhara, 1988), suggesting that some genes involved in spikelet formation might also function in root development. The Arabidopsis ortholog of bd1 is PUCHI, which is required for the restriction of cell division to specific cells during the early stages of lateral root primordium formation (Hirota et al., 2007). Like bd1, PUCHI is also involved in specifying the architecture of the inflorescence (Karim et al., 2009). The wild-type-specific expression of bd1 in coleoptilar nodes at all three developmental stages and the role of its ortholog PUCHI during root meristem development in Arabidopsis suggest a role of bd1 in the formation of crown roots in maize.
Expression Patterns of Phytohormone-Related Genes during Coleoptilar Node Formation
Auxin maxima are required for root primordia formation and outgrowth of root tips (Péret et al., 2009; Jansen et al., 2012). The rtcs gene is an auxin-inducible early auxin-responsive gene that controls seminal and shoot-borne root formation (Taramino et al., 2007). Similarly, the maize rum1 gene encodes an auxin-inducible early auxin-responsive gene of the Aux/IAA family that is required for seminal and lateral root initiation (von Behrens et al., 2011). In this study, the maize Aux/IAA genes iaa9 and iaa15 were preferentially expressed in wild-type t4 coleoptilar nodes, which might imply a role of these genes in shoot-borne root formation.
Furthermore, four genes involved in ethylene biosynthesis (BINCODE 17.5.1) and ethylene signal transduction (BINCODE 17.5.2) are preferentially expressed in wild-type versus rtcs coleoptilar nodes. It has been demonstrated that increased levels of ethylene induce cell death (He et al., 1996a, 1996b). Hence, these genes might be required to facilitate the outgrowth of shoot-borne roots by triggering cell death of cortical cells that have to be penetrated during shoot-borne root emergence. Finally, three genes involved in abscisic acid (ABA) synthesis/degradation (BINCODE 17.1.1) were differentially expressed between WTt0 and rtcst0. ABA inhibits lateral root initiation, while the degradation of ABA promotes lateral root formation. ABA might have a similar function in shoot-borne root initiation.
Distribution of LBD Promoter Motifs and in Vitro Interaction of RTCS with Putative Downstream Target Genes
Thus far, only little is known about direct targets of RTCS. It has been demonstrated that LBD proteins can bind specifically to the 5′-GCGGCG-3′ LBD consensus sequence motif in the promoter of target genes (Husbands et al., 2007). It has been demonstrated that RTCS can bind to the LBD promoter element of arf34 that is located 277 bp upstream of the ATG start codon (Majer et al., 2012). Among the differentially expressed genes identified in this study, 100 of 828 contained at least one LBD motif in their 1-kb promoter. Among those, 65 were preferentially expressed in wild-type and 35 in rtcs coleoptilar nodes. This implies that RTCS can activate or repress downstream genes. Cotransfection assays using RTCS-GAL4 DNA-binding domain fusion proteins as effector and a GAL4-binding site fused to a luciferase reporter suggested that RTCS acts as a transcriptional activator (Majer et al., 2012). However, recently, it was also demonstrated that RTCS can repress the expression of target genes such as arf16 (C. Xu, personal communication). The ability to switch the transcriptional mode of action between activation and repression is known from transcription factors in other plants, such as the repressor WUSCHEL in Arabidopsis, which becomes an activator only in the flower, where it induces AGAMOUS expression (Ikeda et al., 2009). In tomato (Solanum lycopersicum), the activator PTI4 converts to a repressor in a repressosome complex with Silencer Element Binding Factor (González-Lamothe et al., 2008). For AtLOB, reduced DNA-binding affinity was shown in the presence of bHLH048 (Husbands et al., 2007), which could allow the gradual modification of the expression level of LOB target genes. Activation or repression of downstream genes by RTCS could be controlled by a yet unknown cofactor similar to bHLH048 in Arabidopsis.
Differentially expressed genes containing LBD motifs displayed a significantly lower abundance than was expected from the genomic distribution of these promoter motifs (Supplemental Table S5). Genes containing LBD motifs are often transcription factors such as arf34 (Majer et al., 2012). Transcription factors typically display low expression levels that are difficult to capture by microarray analyses due to their weak expression. Moreover, microarray analyses do not only detect direct targets of the mutated genes but also genes that act farther downstream. The fact that the majority of genes differentially expressed between the wild type and rtcs do not contain LBD motifs in their promoters can most likely be explained by the fact that LBD genes are mainly regulatory genes that are expressed at low levels that might not allow identifying them as differentially expressed in microarray experiments. Moreover, these results might indicate that most differentially expressed genes are not direct targets of RTCS but act farther downstream. The putative direct RTCS targets containing LBD motifs are involved in a wide variety of cellular processes. However, none of the MapMan categories except miscellaneous was overrepresented among the differentially expressed genes. For four of these putative direct target promoters, binding of RTCS to a LBD motif has been demonstrated. Previously, RTCS binding to the promoter of arf34 has been demonstrated with the same approach (Majer et al., 2012). Nevertheless, although all genes containing LBD motifs in their promoters are putative RTCS targets, this study can provide only initial clues on such RTCS targets, which need to be studied in more detail by genetic analyses in the future.
In summary, comprehensive microarray analyses based on detailed histological analyses revealed RTCS-dependent regulation of gene expression during the early stages of crown root formation in the coleoptilar node. These results provide, to our knowledge, first insights into the molecular framework associated with crown root formation in maize and a rich source of candidate genes for future genetic analyses of shoot-borne root initiation in maize.
MATERIALS AND METHODS
Plant Materials
Coleoptilar nodes of homozygous wild-type and rtcs maize (Zea mays) were isolated from seedlings germinated in paper rolls in a plant growth chamber at 26°C with a 16-h-light/8-h-dark program at 60% humidity. Homozygous wild-type and mutant plants for these analyses were obtained from phenotypes that were backcrossed more than seven times in the B73 background. This ensured that wild-type and mutant plants were genetically as closely related as possible. Coleoptilar nodes of wild-type and rtcs seedlings were manually dissected from seedlings of homozygous wild-type and rtcs plants 0 d (t0), 2 d (t2), and 4 d (t4) after the appearance of these structures and frozen in liquid nitrogen immediately after isolation. Each coleoptilar node RNA sample (biological replicate) was obtained from a pool of approximately 10 coleoptilar nodes.
Histological Analyses of Coleoptilar Nodes
Coleoptilar nodes were fixed in 4% (w/v) paraformaldehyde for 12 h at 4°C as described previously (Hochholdinger and Feix, 1998). Sections of 12 µm were prepared with a Leica 2035 biocut microtome. Staining of deparaffinized specimens with Safranin O (AppliChem) and Fast Green (Sigma-Aldrich) was performed as described previously (Hetz et al., 1996). Stained coleoptilar node sections were studied using a Zeiss Axioskop HBO 100W/2 microscope and documented with a Canon Powershot G2 digital camera.
RNA Isolation
For microarray experiments, total RNA was isolated from approximately 500 mg of frozen coleoptilar nodes per biological replicate via the RNeasy Plant Mini Kit (Qiagen). For all samples, poly(A) RNA was isolated using the Illustra mRNA Purification Kit (GE Biosciences) and analyzed on Agilent’s Bioanalyzer to check for degradation and to determine concentration. For qRT-PCR, total RNA was isolated from approximately 500 mg of frozen coleoptilar nodes per biological replicate with peqGOLD TriFast reagent (Peqlab). Genomic DNA was removed in both instances by incubating the RNA samples with RNase-free DNase I (Fermentas). All samples were tested for genomic contamination by PCR with primers deduced from a maize actin gene (GenBank accession no. AY104722).
Microarray Hybridization
Microarray analyses were conducted with Pioneer’s custom-made 2x 105 k Agilent 60-mer oligonucleotide maize microarray chips (Agilent Technologies) containing 103,680 microarray features (Gene Expression Omnibus platform GPL10837; Hayes et al., 2010) Since all oligonucleotide sequences were based on DuPont proprietary sequences, these oligonucleotide sequences were BLASTed on an in-house BLAST server against the maize reference complementary DNA (cDNA) data set ZmB73_4a.53_working_cdna (www.maizesequence.org; cutoff level, E ≤ 10−10). This data set represents sequences of 134,099 transcripts. Overall, 65,646 of the proprietary sequences corresponded to cDNA sequences present in the public ZmB73_4a.53_working_cdna database. Only these sequences were included in further analyses. The microarray accessions of these 65,646 transcripts and their corresponding ZmB73_4a.53_working_cdna accessions are listed in Supplemental Table S1. The 65,646 microarray features covered by ZmB73_4a.53_working_cdna represent 37,073 unique transcripts encoded by 31,355 unique high-confidence gene models. Hence, on average, each gene is represented by approximately 1.2 distinct transcripts (e.g. splice variants) and each unique gene model is represented on average by approximately 2.1 features on the microarray chip. The current version of the maize genome annotation postulated 39,656 distinct genes (4a.53_v2; maizesequence.org). Hence, the used microarray chip covers approximately 79% of all high-confidence maize genes.
Cy3-labeled complementary RNA (cRNA) was synthesized from 100 ng of poly(A) RNA using the Quick Amp Labeling Kit One Color (Agilent), which generates fluorescent cRNA by simultaneously amplifying target material and incorporating Cy3-labeled dCTP via T7 polymerase. For each of the three developmental stages and two genotypes, four independent Cy3-labeled cRNA samples were prepared (biological replicates). Hence, 24 Cy3-labeled cRNAs were subsequently hybridized in “one-color mode” to 24 arrays using Agilent’s Gene Expression Hybridization Kit for 17 h at 65°C according to the manufacturer’s protocol. After hybridization, microarrays were washed twice with Agilent’s Gene Expression Wash Buffer 1 and 2 for 1 min each at room temperature and 37°C, respectively, followed by a brief wash in acetonitrile. Scanning of arrays was performed with Agilent’s G2505B DNA Microarray Scanner at 5-μm resolution in the single-path scanning mode of the green dye channel at two laser settings (10% and 100% photomultiplier tube) using Agilent’s Scan Control software, version A.7.0.1. Microarray data of all 24 hybridizations have been deposited as Gene Expression Omnibus series GSE25467.
Microarray Data Normalization, Transformation, and Calculation of Linear Contrasts
The two microarray images were combined, and features were extracted with Agilent’s Feature Extraction 9.5.3 Image Analysis Software. Control features and flagged microarray features were excluded, and the top and bottom 10% of the data were trimmed. Subsequently, the data were normalized via the mean intensity feature of the Rosetta Resolver program (Rosetta Biosoftware). A P value threshold of 0.01 was applied, which was calculated with Agilent’s single-color error model. After normalization, the linear model log2(yij) = ai + eij was fitted for each microarray feature, where log2(yij) is the log2-transformed value of the normalized gene expression level of the jth replicate of genotype/stage combination i, ai is the fixed effect of stage/genotype combination i, and eij is the corresponding residual, assumed to be independently and identically approximately N(0,σ2).
Three pairwise linear contrasts of wild type versus rtcs comparisons (WTt0 versus rtcst0, WTt2 versus rtcst2, and WTt4 versus rtcst4) were tested. To borrow strength across microarray features, we used the empirical Bayes approach of Smyth (2004). Using the R package limma (for linear models for microarray data) for the empirical Bayes approach, moderated P values were calculated for pairwise comparisons. For each contrast, a list of moderated P values over microarray features was then corrected for multiple testing using the positive false discovery rate, which is measured in terms of q values that were then used to identify differentially expressed genes while controlling the false discovery rate at 5% (Storey, 2002; Storey and Tibshirani, 2003).
qRT-PCR
For each of the two genotypes, cDNA was synthesized from 250 ng of total RNA of three independent biological replicates of the three developmental stages using the qScript cDNA SuperMix (Quanta Biosciences). qRT-PCR was performed with three technical replicates per each of the three biological replicates using MESA Green qPCR Mastermix Plus for SYBR Assay No ROX (Eurogentech) and 8 µL of total reaction volume per sample in the CFX384 Real-Time PCR Detection System (Bio-Rad) with PCR FramePlate 384 plates (VWR). Efficiency for all primers was tested by a dilution series (1, 1:2, 1:4, 1:8, 1:16, 1:32, 1:64, and 1:128). Transcript levels were analyzed for all genes with respect to the cDNA level of the H chain of the housekeeping gene myosin (GenBank accession no. 486090G09.x1), which was previously used as a qRT-PCR standard (Dembinsky et al., 2007). Differential expression of each gene was tested using Student’s t test (fold change ≥ 2, P ≤ 0.05).
Annotation of Differentially Accumulated Transcripts via Mercator
All 65,646 microarray features that corresponded to accessions from the maize cDNA database ZmB73_4a.53_working_cdna were functionally annotated via the automated Mercator pipeline (http://www.gabipd.org/biotools/mercator/). The Mercator output files were subsequently used to generate overview maps for differentially expressed genes in MapMan (http://www.gabipd.org/projects/MapMan/). To determine if specific functional groups are overrepresented among the differentially expressed genes with reference to all genes represented on the microarray chip, the expected number of genes for each functional category was calculated based on the distribution of functional categories among all genes on the chip. To determine if significantly more or fewer genes than expected were detected for each individual category, a χ2 test for independence with Yates’ continuity correction was performed.
Distribution of LBD Motifs
The frequency distribution of the number of LBD motifs in the 1-kb region immediately upstream of the ATG start codon between differentially expressed genes and the remaining genes among all 39,656 genes was compared. The null hypothesis that both distributions are identical was tested by a test of independence in a 2 × 2 contingency table of two types of genes (differentially expressed and not differentially expressed) and a dichotomized classification of the LBD counts (zero and one or more) by an asymptotic χ2 test. Due to the small number of observed counts for genes with a larger number of LBD motifs, we performed an exact test using the FREQ procedure of the SAS System. The sample size of 39,656 genes was too large for a complete enumeration by the network algorithm of Mehta (1983). Thus, we used the Monte Carlo algorithm of Agresti et al. (1979) based on n = 10,000 simulation runs to approximate the exact P value of the likelihood ratio χ2 test statistic.
EMSA
N-terminal GST fused to the open reading frame of rtcs was introduced into the vector pGEX-6P-1 (Smyth, 2004) and expressed in Escherichia coli BL21-DE3 cells. Because of inclusion body formation, the crude extract was isolated in the presence of 0.25% sarcosyl (Frangioni and Neel, 1993). EMSAs were performed as described in Promega technical bulletin TB110 (www.promega.com/tbs/tb110/tb110.pdf). The target sequences used for the DNA/protein-binding reaction (Supplemental Table S6) were labeled with [γ-32P]ATP (Hartmann Analytic) and T4 polynucleotide kinase (Fermentas). Crude protein extract (20 μg) containing recombinant GST-RTCS fusion proteins was incubated with [γ-32P]ATP-labeled DNA fragment and 1 μg of poly(dI-dC) in 10× buffer (100 mm Tris, pH 7.5, 500 mm NaCl, 10 mm EDTA, and 10 mm dithiothreitol) in a total volume of 40 μL. The reaction products were analyzed on 4% nondenaturing polyacrylamide gels. The specificity of GST-RTCS binding was controlled by using either 50-fold excess of specific competitor (unlabeled target sequence summarized in Supplemental Table S6) or sonicated herring sperm DNA (Promega) as unspecific competitor DNA that provided a mixture of distinct DNA motifs of the appropriate size.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table S1. A total of 65,646 microarray features mapped to transcript accessions of the maize B73 working cDNA set (version 4a.53).
Supplemental Table S2. Microarray features differentially expressed between wild-type and mutant rtcs with MapMan annotations generated by Mercator.
Supplemental Table S3. qRT-PCR confirmation of differentially expressed genes between the wild type and rtcs.
Supplemental Table S4. Differentially expressed microarray features from Supplemental Table S2 located within genes that contain one or more LBD motifs within an interval 1 kb upstream of their ATG start codons.
Supplemental Table S5. Distribution of LBD motifs among all high-confidence genes of the maize filtered gene set and the differentially expressed genes identified in this study.
Supplemental Table S6. Oligonucleotide primer sequences used for qRT-PCR and EMSA experiments.
Acknowledgments
We thank Dieter Steinmetz (University of Tuebingen) for help with Perl and visual basic for applications scripting, Angela Dressel (University of Tuebingen) for excellent technical assistance, Kevin Hayes (Pioneer Hi-Bred) for support with microarray annotation, and Marc Lohse for generating the mapping of ZmB73_4a.53_working_cdna using the Mercator pipeline.
Glossary
- qRT
quantitative real-time
- EMSA
electrophoretic mobility shift assay
- GST
glutathione S-transferase
- ABA
abscisic acid
- cDNA
complementary DNA
- cRNA
complementary RNA
References
- Agresti A, Wackerly D, Boyett JM. (1979) Exact conditional tests for cross-classifications: approximation of attained significance levels. Psychometrika 44: 75–83 [Google Scholar]
- Aida M, Beis D, Heidstra R, Willemsen V, Blilou I, Galinha C, Nussaume L, Noh YS, Amasino R, Scheres B. (2004) The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell 119: 109–120 [DOI] [PubMed] [Google Scholar]
- Alexandrov NN, Brover VV, Freidin S, Troukhan ME, Tatarinova TV, Zhang HY, Swaller TJ, Lu YP, Bouck J, Flavell RB, et al. (2009) Insights into corn genes derived from large-scale cDNA sequencing. Plant Mol Biol 69: 179–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortiri E, Chuck G, Vollbrecht E, Rocheford T, Martienssen R, Hake S. (2006) ramosa2 encodes a LATERAL ORGAN BOUNDARY domain protein that determines the fate of stem cells in branch meristems of maize. Plant Cell 18: 574–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalfun A, Jr, Franken J, Mes JJ, Marsch-Martinez N, Pereira A, Angenent GC. (2005) ASYMMETRIC LEAVES2-LIKE1 gene, a member of the AS2/LOB family, controls proximal-distal patterning in Arabidopsis petals. Plant Mol Biol 57: 559–575 [DOI] [PubMed] [Google Scholar]
- Chuck G, Muszynski M, Kellogg E, Hake S, Schmidt RJ. (2002) The control of spikelet meristem identity by the branched silkless1 gene in maize. Science 298: 1238–1241 [DOI] [PubMed] [Google Scholar]
- Dembinsky D, Woll K, Saleem M, Liu Y, Fu Y, Borsuk LA, Lamkemeyer T, Fladerer C, Madlung J, Barbazuk B, et al. (2007) Transcriptomic and proteomic analyses of pericycle cells of the maize primary root. Plant Physiol 145: 575–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans MMS. (2007) The indeterminate gametophyte1 gene of maize encodes a LOB domain protein required for embryo sac and leaf development. Plant Cell 19: 46–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frangioni JV, Neel BG. (1993) Solubilization and purification of enzymatically active glutathione S-transferase (pGEX) fusion proteins. Anal Biochem 210: 179–187 [DOI] [PubMed] [Google Scholar]
- Gardiner J, Schroeder S, Polacco ML, Sanchez-Villeda H, Fang ZW, Morgante M, Landewe T, Fengler K, Useche F, Hanafey M, et al. (2004) Anchoring 9,371 maize expressed sequence tagged unigenes to the bacterial artificial chromosome contig map by two-dimensional overgo hybridization. Plant Physiol 134: 1317–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Lamothe R, Boyle P, Dulude A, Roy V, Lezin-Doumbou C, Kaur GS, Bouarab K, Després C, Brisson N. (2008) The transcriptional activator Pti4 is required for the recruitment of a repressosome nucleated by repressor SEBF at the potato PR-10a gene. Plant Cell 20: 3136–3147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes KR, Beatty M, Meng X, Simmons CR, Habben JE, Danilevskaya ON. (2010) Maize global transcriptomics reveals pervasive leaf diurnal rhythms but rhythms in developing ears are largely limited to the core oscillator. PLoS ONE 5: e12887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He CJ, Finlayson SA, Drew MC, Jordan WR, Morgan PW. (1996a) Ethylene biosynthesis during aerenchyma formation in roots of maize subjected to mechanical impedance and hypoxia. Plant Physiol 112: 1679–1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He CJ, Morgan PW, Drew MC. (1996b) Transduction of an ethylene signal is required for cell death and lysis in the root cortex of maize during aerenchyma formation induced by hypoxia. Plant Physiol 112: 463–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetz W, Hochholdinger F, Schwall M, Feix G. (1996) Isolation and characterization of rtcs, a maize mutant deficient in the formation of nodal roots. Plant J 10: 845–857 [Google Scholar]
- Hirota A, Kato T, Fukaki H, Aida M, Tasaka M. (2007) The auxin-regulated AP2/EREBP gene PUCHI is required for morphogenesis in the early lateral root primordium of Arabidopsis. Plant Cell 19: 2156–2168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochholdinger F (2009) The maize root system: morphology, anatomy, and genetics. In JL Bennetzen, SC Hake, eds, Handbook of Maize: Its Biology. Springer, New York, pp 145–160 [Google Scholar]
- Hochholdinger F, Feix G. (1998) Cyclin expression is completely suppressed at the site of crown root formation in the nodal region of the maize root mutant rtcs. J Plant Physiol 153: 425–429 [Google Scholar]
- Hochholdinger F, Park WJ, Sauer M, Woll K. (2004a) From weeds to crops: genetic analysis of root development in cereals. Trends Plant Sci 9: 42–48 [DOI] [PubMed] [Google Scholar]
- Hochholdinger F, Woll K, Sauer M, Dembinsky D. (2004b) Genetic dissection of root formation in maize (Zea mays L.) reveals root-type specific developmental programmes. Ann Bot (Lond) 93: 359–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoecker N, Keller B, Muthreich N, Chollet D, Descombes P, Piepho HP, Hochholdinger F. (2008) Comparison of maize (Zea mays L.) F1-hybrid and parental inbred line primary root transcriptomes suggests organ-specific patterns of nonadditive gene expression and conserved expression trends. Genetics 179: 1275–1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppe DC, McCully ME, Wenzel CL. (1986) The nodal roots of Zea: their development in relation to structural features of the stem. Can J Bot 64: 2524–2537 [Google Scholar]
- Husakova E, Hochholdinger F, Soukup A. (2013) Lateral root development in the maize (Zea mays) lateral rootless1 mutant. Ann Bot (Lond) 112: 417–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husbands A, Bell EM, Shuai B, Smith HMS, Springer PS. (2007) LATERAL ORGAN BOUNDARIES defines a new family of DNA-binding transcription factors and can interact with specific bHLH proteins. Nucleic Acids Res 35: 6663–6671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda M, Mitsuda N, Ohme-Takagi M. (2009) Arabidopsis WUSCHEL is a bifunctional transcription factor that acts as a repressor in stem cell regulation and as an activator in floral patterning. Plant Cell 21: 3493–3505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inukai Y, Sakamoto T, Ueguchi-Tanaka M, Shibata Y, Gomi K, Umemura I, Hasegawa Y, Ashikari M, Kitano H, Matsuoka M. (2005) crown rootless1, which is essential for crown root formation in rice, is a target of an AUXIN RESPONSE FACTOR in auxin signaling. Plant Cell 17: 1387–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwakawa H, Iwasaki M, Kojima S, Ueno Y, Soma T, Tanaka H, Semiarti E, Machida Y, Machida C. (2007) Expression of the ASYMMETRIC LEAVES2 gene in the adaxial domain of Arabidopsis leaves represses cell proliferation in this domain and is critical for the development of properly expanded leaves. Plant J 51: 173–184 [DOI] [PubMed] [Google Scholar]
- Iwakawa H, Ueno Y, Semiarti E, Onouchi H, Kojima S, Tsukaya H, Hasebe M, Soma T, Ikezaki M, Machida C, et al. (2002) The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana, required for formation of a symmetric flat leaf lamina, encodes a member of a novel family of proteins characterized by cysteine repeats and a leucine zipper. Plant Cell Physiol 43: 467–478 [DOI] [PubMed] [Google Scholar]
- Jansen L, Roberts I, De Rycke R, Beeckman T. (2012) Phloem-associated auxin response maxima determine radial positioning of lateral roots in maize. Philos Trans R Soc Lond B Biol Sci 367: 1525–1533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JW, Pope MN. (1942) Adventitious roots on panicles of rice. J Hered 33: 55–58 [Google Scholar]
- Karim MR, Hirota A, Kwiatkowska D, Tasaka M, Aida M. (2009) A role for Arabidopsis PUCHI in floral meristem identity and bract suppression. Plant Cell 21: 1360–1372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HW, Kim NY, Lee DJ, Kim J. (2009) LBD18/ASL20 regulates lateral root formation in combination with LBD16/ASL18 downstream of ARF7 and ARF19 in Arabidopsis. Plant Physiol 151: 1377–1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li A, Zhang Y, Wu X, Tang W, Wu R, Dai Z, Liu G, Zhang H, Wu C, Chen G, et al. (2008) DH1, a LOB domain-like protein required for glume formation in rice. Plant Mol Biol 66: 491–502 [DOI] [PubMed] [Google Scholar]
- Lin WC, Shuai B, Springer PS. (2003) The Arabidopsis LATERAL ORGAN BOUNDARIES-domain gene ASYMMETRIC LEAVES2 functions in the repression of KNOX gene expression and in adaxial-abaxial patterning. Plant Cell 15: 2241–2252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HJ, Wang SF, Yu XB, Yu J, He XW, Zhang SL, Shou HX, Wu P. (2005) ARL1, a LOB-domain protein required for adventitious root formation in rice. Plant J 43: 47–56 [DOI] [PubMed] [Google Scholar]
- Majer C, Hochholdinger F. (2011) Defining the boundaries: structure and function of LOB domain proteins. Trends Plant Sci 16: 47–52 [DOI] [PubMed] [Google Scholar]
- Majer C, Xu CZ, Berendzen KW, Hochholdinger F. (2012) Molecular interactions of ROOTLESS CONCERNING CROWN AND SEMINAL ROOTS, a LOB domain protein regulating shoot-borne root initiation in maize (Zea mays L.). Philos Trans R Soc Lond B Biol Sci 367: 1542–1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumura Y, Iwakawa H, Machida Y, Machida C. (2009) Characterization of genes in the ASYMMETRIC LEAVES2/LATERAL ORGAN BOUNDARIES (AS2/LOB) family in Arabidopsis thaliana, and functional and molecular comparisons between AS2 and other family members. Plant J 58: 525–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta CRPN. (1983) A network algorithm for performing Fisher’s exact test in r contingency tables. J Am Stat Assoc 78: 427–434 [Google Scholar]
- Nass H, Zuber MS. (1971) Correlation of corn roots in early development to mature root development. Crop Sci 11: 655–658 [Google Scholar]
- Okushima Y, Fukaki H, Onoda M, Theologis A, Tasaka M. (2007) ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. Plant Cell 19: 118–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschold A, Jia Y, Marcon C, Lund S, Larson NB, Yeh CT, Ossowski S, Lanz C, Nettleton D, Schnable PS, et al. (2012) Complementation contributes to transcriptome complexity in maize (Zea mays L.) hybrids relative to their inbred parents. Genome Res 22: 2445–2454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Péret B, De Rybel B, Casimiro I, Benková E, Swarup R, Laplaze L, Beeckman T, Bennett MJ. (2009) Arabidopsis lateral root development: an emerging story. Trends Plant Sci 14: 399–408 [DOI] [PubMed] [Google Scholar]
- Sauer M, Jakob A, Nordheim A, Hochholdinger F. (2006) Proteomic analysis of shoot-borne root initiation in maize (Zea mays L.). Proteomics 6: 2530–2541 [DOI] [PubMed] [Google Scholar]
- Smyth GK. (2004) Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3: Article3 [DOI] [PubMed] [Google Scholar]
- Storey JD. (2002) A direct approach to false discovery rates. J R Stat Soc Ser B Stat Methodol 64: 479–498 [Google Scholar]
- Storey JD, Tibshirani R. (2003) Statistical significance for genomewide studies. Proc Natl Acad Sci USA 100: 9440–9445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi M, Futsuhara J. (1988) A dense panicle mutant producing adventitious roots from spikelets. Rice Genet Newsl 5: 113 [Google Scholar]
- Taramino G, Sauer M, Stauffer JL, Jr, Multani D, Niu XM, Sakai H, Hochholdinger F. (2007) The maize (Zea mays L.) RTCS gene encodes a LOB domain protein that is a key regulator of embryonic seminal and post-embryonic shoot-borne root initiation. Plant J 50: 649–659 [DOI] [PubMed] [Google Scholar]
- von Behrens I, Komatsu M, Zhang YX, Berendzen KW, Niu XM, Sakai H, Taramino G, Hochholdinger F. (2011) rootless with undetectable meristem 1 encodes a monocot-specific AUX/IAA protein that controls embryonic seminal and post-embryonic lateral root initiation in maize. Plant J 66: 341–353 [DOI] [PubMed] [Google Scholar]
- Woll K, Borsuk LA, Stransky H, Nettleton D, Schnable PS, Hochholdinger F. (2005) Isolation, characterization, and pericycle-specific transcriptome analyses of the novel maize lateral and seminal root initiation mutant rum1. Plant Physiol 139: 1255–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Yu XB, Wu P. (2006) Comparison and evolution analysis of two rice subspecies LATERAL ORGAN BOUNDARIES domain gene family and their evolutionary characterization from Arabidopsis. Mol Phylogenet Evol 39: 248–262 [DOI] [PubMed] [Google Scholar]