Expression of xyloglucan arabinofuranosyltransferases, identified from tomato using a comparative genomics approach, rescues growth and mechanical defects of an Arabidopsis mutant deficient for xyloglucan galactosylation.
Abstract
Xyloglucan (XyG) is the dominant hemicellulose present in the primary cell walls of dicotyledonous plants. Unlike Arabidopsis (Arabidopsis thaliana) XyG, which contains galactosyl and fucosyl substituents, tomato (Solanum lycopersicum) XyG contains arabinofuranosyl residues. To investigate the biological function of these differing substituents, we used a functional complementation approach. Candidate glycosyltransferases were identified from tomato by using comparative genomics with known XyG galactosyltransferase genes from Arabidopsis. These candidate genes were expressed in an Arabidopsis mutant lacking XyG galactosylation, and two of them resulted in the production of arabinosylated XyG, a structure not previously found in this plant species. These genes may therefore encode XyG arabinofuranosyltransferases. Moreover, the addition of arabinofuranosyl residues to the XyG of this Arabidopsis mutant rescued a growth and cell wall biomechanics phenotype, demonstrating that the function of XyG in plant growth, development, and mechanics has considerable flexibility in terms of the specific residues in the side chains. These experiments also highlight the potential of reengineering the sugar substituents on plant wall polysaccharides without compromising growth or viability.
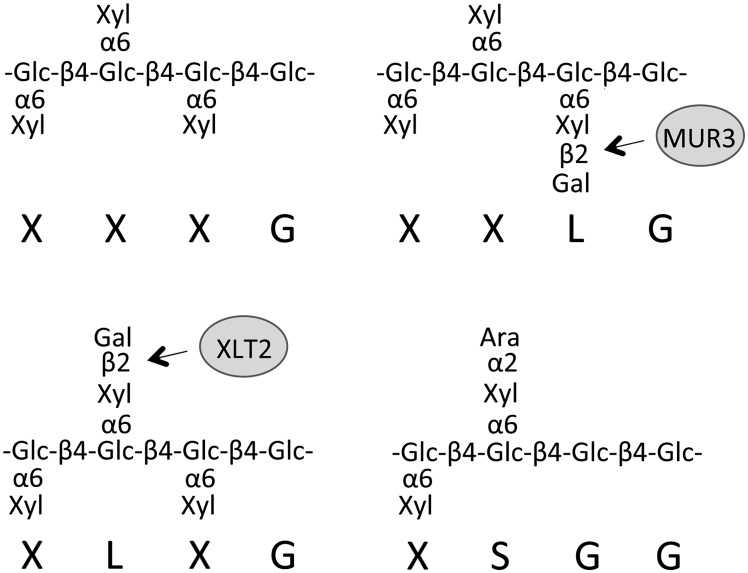
The cell wall of higher plants represents a composite material consisting of various polymers including cellulose, hemicellulose, lignin, pectin, and glycoproteins (Somerville et al., 2004). The quantity and fine structure of each of these components varies based on the tissue type and plant species (Pauly and Keegstra, 2008). One of the major components of the dicot primary wall (the wall of growing cells) is the hemicellulose xyloglucan (XyG), whose structure and biosynthesis are relatively well described (Zabotina, 2012). The glycan backbone of XyG consists of β-1,4-linked glucosyl residues, which are substituted with a regular pattern of xylosyl residues that can be further decorated with a diverse array of carbohydrate and noncarbohydrate substituents. A one-letter code nomenclature has been developed to specify the substituents of a particular backbone glucosyl residue (Fry et al., 1993). An unsubstituted Glc residue is depicted as G while a Glc substituted with a xylosyl residue is depicted as X. Further substitution of the Xyl with a β-galactosyl or α-arabinofuranosyl-residue is abbreviated as L or S, respectively (Fig. 1). In addition, an L side chain may contain an α-fucosyl moiety on the Gal (abbreviated F) or an acetyl group (underlined L). More than 10 additional side chain structures have been identified in various plant species (Hantus et al., 1997; Jia et al., 2003; Ray et al., 2004; Peña et al., 2008, 2012).
Figure 1.
Xyloglucan motifs present in the walls of Arabidopsis (XXXG, XXLG, and XLXG) and tomato (XSGG) with nomenclature indicated below the structure. AtMUR3 and AtXLT2 are galactosyltransferases required to produce XXLG and XLXG, respectively. Glc, d-Glucopyranose; Xyl, d-xylopyranose; Gal, d-galactopyranose; Ara, l-arabinofuranose.
The analysis of XyG structure is facilitated by the availability of a XyG-specific endoglucanase (XEG) that can release XyG oligosaccharides from plant cell wall preparations (Pauly et al., 1999). When XyG is enzymatically released from the walls of the plant model species Arabidopsis (Arabidopsis thaliana), the oligosaccharides XXXG, XXLG, XXLG, XXFG, XXFG, XLFG, and XLFG are observed (Scheller and Ulvskov, 2010), the structures of several of which are shown in Figure 1. Arabinosylated side chains (S) have not been observed in Arabidopsis walls but are abundant in Solanaceous species such as tomato (Solanum lycopersicum). Tomato XyG consists primarily of the subunits LSGG, XSGG, LXGG, LLGG, and XXGG (Jia et al., 2003, 2005). Unlike the “XXXG” type motif in Arabidopsis, XyG in tomato is less xylosylated, with a repeating “XXGG” type motif. In addition, the glucan backbone of tomato XyG is O-acetylated, a modification that in Arabidopsis is only observed on the side chain galactosyl moiety. The functional significance and genetic basis of these structural differences are not understood.
Numerous genes, mainly identified from Arabidopsis, are known to be involved in the biosynthesis of XyG (Pauly et al., 2013). These include a glucan synthase, a member of the Cellulose Synthase-Like C gene family (Cocuron et al., 2007), several XyG xylosyltransferases (XXTs; Faik et al., 2002; Cavalier and Keegstra, 2006), the galactosyltransferases MURUS3 (MUR3; Madson et al., 2003) and XyG Galactose Transferase at Position 2 (XLT2; Jensen et al., 2012), a XyG-specific galacturonosyltransferase (XUT1; Peña et al., 2012), the fucosyltransferase MUR2 (Perrin et al., 1999; Vanzin et al., 2002), and the XyG O-acetyltransferases Altered XyG4 (AXY4) and AXY4-Like (AXY4L; Gille et al., 2011). The Cellulose Synthase-Like C from nasturtium (Tropaeolum majus; Cocuron et al., 2007), where XyG is produced as a seed storage polymer, and MUR3 from eucalyptus (Eucalyptus grandis; Lopes et al., 2010) have both been investigated and appear to have specificities similar to their Arabidopsis orthologs. Glycosyltransferases (GTs) with novel specificities required for the diversity of XyG substitution found in various non-Arabidopsis species, including a XyG arabinosyltransferase, have not been identified to date.
XyG figures prominently in many models of the plant cell wall, where it is thought to cross link cellulose microfibrils and have a mechanistic role in cell elongation (Somerville et al., 2004; Hayashi and Kaida, 2011). However, the recent discovery that an Arabidopsis xxt1 xxt2 double mutant lacks detectable XyG but only has relatively minor growth phenotypes questions the structural significance of this polysaccharide (Cavalier et al., 2008; Park and Cosgrove, 2012b). XyG substitution has been shown to influence polymer solubility and binding affinity in vitro, with the enzymatic removal of side chains leading to decreased polymer solubility (Sims et al., 1998; Lima et al., 2004). However, mutants deficient for MUR2, XLT2, or AXY4 show minor, if any, growth phenotypes under laboratory conditions (Vanzin et al., 2002; Gille et al., 2011; Jensen et al., 2012). A point mutant in MUR3 (mur3.1) shows minor growth phenotypes (Madson et al., 2003), while a transfer DNA mutant in MUR3 shows impaired growth and altered Golgi structure (Tamura et al., 2005). The difference between these alleles was attributed to a role of the MUR3 protein interacting with actin to help organize Golgi structure independent from its function as a GT (Tamura et al., 2005). XyG oligosaccharides, which can be liberated by endogenous or exogenous glycosyl hydrolases, have been suggested to have a role in signaling (Aldington et al., 1991), though the biological significance of this is unclear and a specific pathway has not been identified. A complimentary approach to using mutant lines lacking certain XyG substitutions to investigate the function of XyG substitution would be to introduce exogenous side chain structures by the expression of XyG biosynthetic genes from other species. This functional complementation approach requires the identification of the genes responsible for exogenous substitution patterns.
The MUR3, XLT2, and XUT1 genes are in the same subclade of the inverting GT family 47 (Li et al., 2004). These three transferases all add β-glycosyl groups to the O2-position of a xylosyl group on XyG but differ in donor specificity, with MUR3 and XLT2 utilizing UDP-Gal and XUT1 utilizing UDP-GalA. The diversity of donor substrate specificity present in this subclade of GTs suggests that similar enzymes may represent good candidate genes for unidentified XyG GTs responsible for XyG side chain diversity in other species.
Here, we report the identification of several tomato genes involved in XyG biosynthesis. Candidate genes were constitutively expressed in the Arabidopsis mur3.1 xlt2 double mutant, which contains mostly nonsubstituted XyG, for functional characterization. Two putative XyG arabinofuranosyltransferases were identified, and the evolutionary history of these genes was investigated using phylogenetics. The expression of these genes rescued the growth and petiole extensibility phenotypes of the mutant, demonstrating partial functional redundancy of XyG galactosylation and arabinosylation.
RESULTS
Phylogenetic Analysis
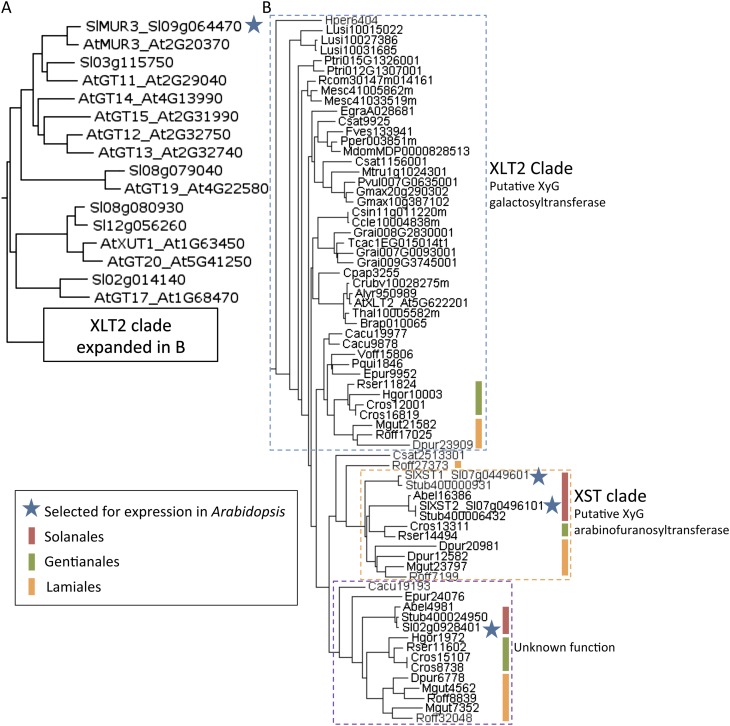
A phylogenetic tree was constructed of the GT family 47 subclade containing MUR3 and XLT2 (Li et al., 2004) using available angiosperm protein sequences from Phytozome and the Medical Plant Genomics Resource (Fig. 2). Nine tomato genes were identified, one of which, Sl09g064470, was most similar to AtMUR3, whereas three (Sl02g092840, Sl07g044960, and Sl07g049610) were most similar to AtXLT2. The four genes were cloned and transformed into the Arabidopsis mur3.1 xlt2 double mutant for ubiquitous expression. Quantitative reverse transcription-PCR was performed on leaf tissue harvested from the T2 generation and indicated that all genes were successfully expressed in the lines analyzed (Supplemental Fig. S1).
Figure 2.
Phylogenetic analysis of the MUR3 and XLT2 subclade of GT family 47. A, Maximum-likelihood tree of Arabidopsis (At) and tomato (Sl) protein sequences. B, The XLT2 clade was expanded with sequences from additional species. The protein sequences are labeled with abbreviations for species (see below) and the gene model (Phytozome) or EST identifier (Medical Plants Genomics Resource). Abel, Atropa belladonna; Alyr, Arabidopsis lyrata; At, Arabidopsis; Brap, Brassica rapa; Cacu, Camptotheca acuminata; Ccle, Citrus clementina; Cpap, Carica papaya; Cros, Catharanthus roseus; Crub, Capsella rubella; Csat, Cannabis sativa; Csin, Citrus sinensis; Dpur, Digitalis purpurea; Dvil, Dioscorea villosa; Egra, Eucalyptus grandis; Epur, Echinacea purpurea; Fves, Fragaria vesca; Gbil, Ginkgo biloba; Gmax, Glycine max; Grai, Gossypium raimondii; Hgor, Hoodia gordonii; Hper, Hypericum perforatum; Lusi, Linum usitatissimum; Mdom, Malus domestica; Mesc, Manihot esculenta; Mgut, Mimulus guttatus; Mtru, Medicago truncatula; Pper, Prunus persica; Pqui, Panax quinquefolius; Ptri, Populus trichocarpa; Pvul, Phaseolus vulgaris; Rcom, Ricinus communis; Roff, Rosmarinus officinalis; Rser, Rauvolfia serpentina; Sl, tomato; Stub, Solanum tuberosum; Tcac, Theobroma cacao; Thal, Thellungiella halophila; Voff, Valeriana officinalis.
Effect on XyG structure
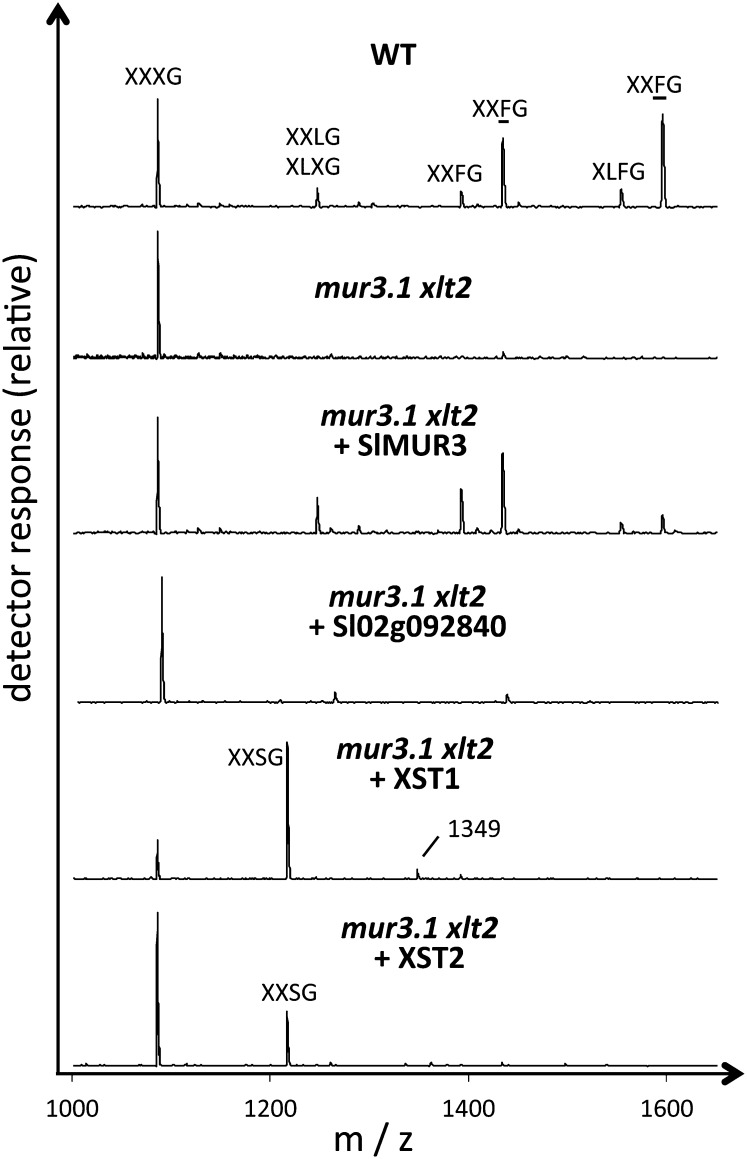
The XyG structure in several independently transformed lines for each transgene was analyzed by XyG oligosaccharide profiling utilizing matrix-assisted laser-desorption ionization-time-of-flight (MALDI-TOF) mass spectrometry (MS) and high-performance anion-exchange chromatography with pulsed amperometric detection (HPAEC-PAD; Fig. 3; Supplemental Figs. S2 and S3; Lerouxel et al., 2002). As previously described, the oligosaccharides detected in the mur3.1 xlt2 mutant consisted primarily of XXXG (Jensen et al., 2012). Expression of Sl09g064470 in this mutant resulted in the generation of additional oligosaccharides consistent by mass and retention time with XXLG, XXFG, and minor amounts of XLFG, as well as their O-acetylated forms (Fig. 3; Supplemental Fig. S3). This XyG structure resembles that of the Arabidopsis AtXLT2 single mutant, indicating that Sl09g064470 is functionally equivalent to AtMUR3 and mediates the galactosylation of XXXG on the third xylosyl residue from the nonreducing end. Sl09g064470 was therefore named SlMUR3.
Figure 3.
XyG oligosaccharide profile by MALDI-TOF MS from leaf tissue of the Arabidopsis wild type (WT; ecotype Columbia), the double mutant mur3.1 xlt2, and transgenic lines of mur3.1 xlt2 expressing the indicated genes from tomato. Assignment of oligosaccharide structures by one-letter code (Fig. 1) as described in the text.
Expression of Sl02g092840 in the Arabidopsis mur3.1 xlt2 mutant did not change the XyG oligosaccharide profile (Fig. 3; Supplemental Fig. S3), suggesting that Sl02g092840 is either inactive, not involved in XyG biosynthesis or the appropriate donor or acceptor substrates are not present in the Arabidopsis double mutant.
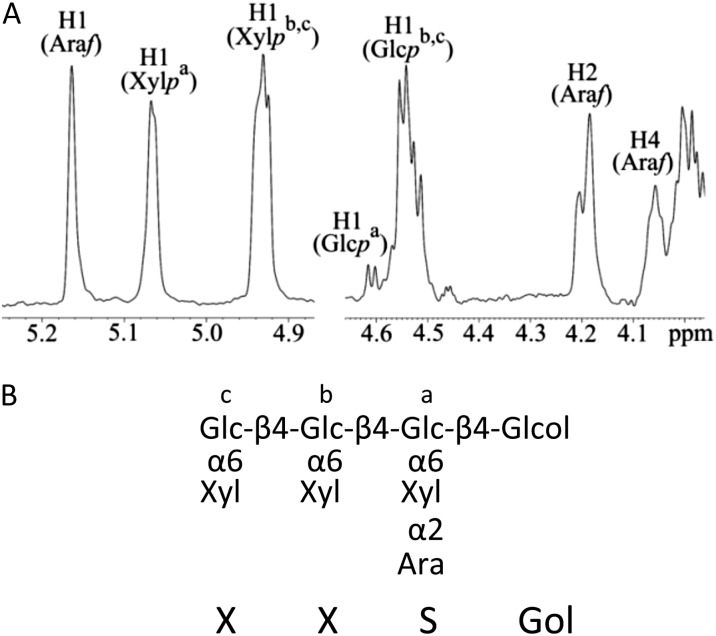
Expression of either Sl07g044960 or Sl07g049610 resulted in the appearance of an XyG oligosaccharide with a mass-to-charge ratio (m/z) of 1,217 (Fig. 3), consistent with an oligosaccharide containing four pentoses and four hexoses. Linkage analysis of this oligosaccharide (purified by HPAEC) revealed the presence of terminal arabinofuranose, possibly attached to the O2-position of a xylosyl residue, in addition to linkages associated with the XXXG oligosaccharide (Supplemental Fig. S4). To determine which xylosyl residue had the attached arabinofuranose, the purified oligosaccharide was digested with the oligoxyloglucan reducing end-specific xyloglucanbiohydrolase (OREX) enzyme (Bauer et al., 2005), which can hydrolyze XXXG to the cellobiose backbone subunits. This digest resulted in the disappearance of the 1,217 m/z ion and the appearance of a 629 m/z ion (Supplemental Fig. S5), representing an oligosaccharide consisting of two pentoses and two hexoses. This data are consistent with an OREX digestion of XXSG to XX and SG. The XyG oligomer XXSG had been previously isolated and characterized from olive (Olea europaea) pulp (Vierhuis et al., 2001). HPAEC analysis of XXSG prepared from olive pulp has the same retention time and mass as the unusual XyG oligosaccharide found in the mur3.1 xtl2 Arabidopsis mutant expressing Sl07g044960 (Supplemental Fig. S6). In addition, 1H-nuclear magnetic resonance (NMR) was performed on the purified unusual oligosaccharide, and the observed chemical shifts were identical to those of the previously published NMR spectrum for XXSG derived from olive pulp (Fig. 4; Supplemental Fig. S7; Vierhuis et al., 2001). Hence, the addition of either Sl07g044960 or Sl07g049610 to the mur3.1 xlt2 double mutant leads to arabinosylation of XyG in Arabidopsis. The genes were hence named XyG “S”-side chain transferase1 (XST1) and XST2, respectively. The abundance of XXSG was higher in the XST1 lines compared with the XST2 lines (Supplemental Figs. S2 and S3). The XST1 lines also had a minor ion in the MALDI-TOF spectra with an m/z of 1,349, which would be consistent with an XXSG oligosaccharide containing an additional pentose (Fig. 3). Future experiments are needed to establish the nature and position of this additional pentose.
Figure 4.
Structure of the novel oligosaccharide. A, 1H-NMR spectrum of the oligosaccharide with an m/z of 1,217 produced in the XST1 overexpression line. B, Proposed structure of the oligosaccharide (reduced to alditol prior to NMR). Superscripts in the spectrum in A refer to the glycosyl units in B. See Supplemental Figure S5 for detailed assignments. Glc, d-Glucopyranose; Xyl, d-xylopyranose; Gal, d-galactopyranose; Ara, l-arabinofuranose.
Effect on Plant Morphology and Cell Wall Biomechanics
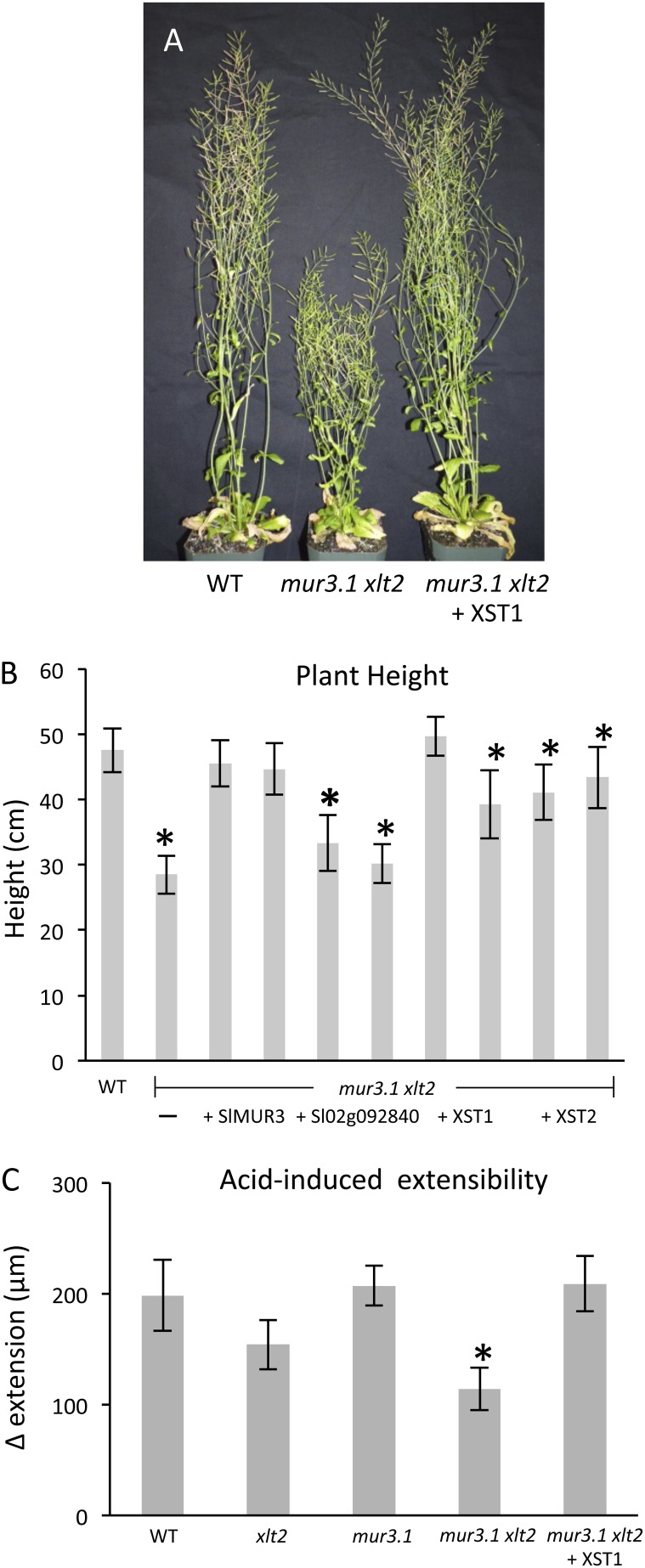
The Arabidopsis mur3.1 xlt2 double mutant exhibits dwarfism compared with wild-type or single mutant plants (Jensen et al., 2012). To investigate how this growth phenotype is affected by alteration of XyG structure the heights of wild-type and mur3.1 xlt2 plants as well as plants from two independently transformed lines for each heterologously expressed tomato gene were measured. Expression of Sl02g092840, which did not affect the XyG structure, resulted in the dwarfism phenotype being retained, while the expression of SlMUR3, XST1, or XST2 resulted in an increase in plant height compared with the nontransgenic double mutant (P ≤ 0.02; Fig. 5; Supplemental Fig. S8). Despite a significant increase, the average heights of plants from XST1- and XST2-expressing lines were less than that of the wild-type plants for three of the four lines examined. This variability may be the result of differential gene expression between the transgenic lines as observed by quantitative PCR analysis (Supplemental Fig. S1) and differing quantities of XyG substitution (Supplemental Fig. S2). Expression of SlMUR3, resulting in a galactosylated, fucosylated XyG, restored plant height completely in both lines examined (Fig. 5B).
Figure 5.
Plant phenotypes and biomechanics. A, Representative 7-week-old Arabidopsis plants for the indicated lines (see Supplemental Fig. S6 for images of all lines). B, Average plant height at 7 weeks with two independent lines for each construct. Error bars indicate sd (n ≥ 4). C, Acid-induced extensibility of leaf petioles under constant force for indicated lines. Error bars show se (n ≥ 9). Asterisk indicates statistically significant difference from the wild type (WT; P < 0.05).
Biomechanical tests on cell wall specimens from leaf petioles of the various lines showed the mur3.1 xlt2 double mutant to be less extensible as assessed by acid-induced creep, which is mediated by the wall-loosening protein α-expansin (Cosgrove, 2000; Sampedro and Cosgrove, 2005; Park and Cosgrove, 2012a). Expression of XST1 restored acid-induced cell wall creep back to wild-type level (Fig. 5C). However, this reduction in extensibility in the mur3.1 xlt2 double mutant and its reversal by XST1 expression were evidently not due to simple changes in mechanical compliance as assessed by stress strain analyses, which was the same in both lines (Supplemental Fig. S9); instead, XyG substitution appeared to be important for the wall-loosening action of endogenous α-expansins.
DISCUSSION
SlMUR3: A Tomato XyG Galactosyltransferase
In vitro activity assays have previously demonstrated that the AtMUR3 protein acts as a XyG galactosyltransferase specific for the third position of the XXXG motif (Madson et al., 2003). Putative orthologs to AtMUR3 can be found in many angiosperm species including nasturtium (Jensen et al., 2012) and eucalyptus, where the MUR3 gene was cloned and shown to functionally complement the Arabidopsis mur3 mutant (Lopes et al., 2010). Unlike Arabidopsis, eucalyptus, and nasturtium, which have XXXG type XyG, tomato XyG contains primarily the XXGG motif. It was therefore unknown if tomato would require a gene with orthologous function to MUR3 for XyG biosynthesis. Through the genetic complementation approach described above, an ortholog to AtMUR3 was identified in tomato (SlMUR3) representing a XyG galactosyltransferase with a similar specificity as AtMUR3. Interestingly, while the XyG structure of the Arabidopsis lines expressing SlMUR3 consisted primarily of XXXG, XXLG, XXFG, and XXFG (consistent with a plant with a defect only in the XLT2), minor amounts of XLFG and XLFG were detected. This may indicate that SlMUR3 also mediates galactosylation of the second position of the XXXG motif, which has not been observed for the AtMUR3 gene. While AtMUR3 has been shown to be active on XXXG, it is unclear if it can act on XXGG. That tomato has retained a gene with MUR3-like activity despite apparently lacking the XXXG motif suggests that SlMUR3 may be able to act on the XXGG motif to form XLGG, a structure that is found in tomato. Alternatively, the XXXG motif may be transiently present in tomato, and a xylosidase may process it to the XXGG form. Future work to test MUR3 activity on various substrates in vitro or a crystal structure of the MUR3 protein bound to substrate would help to reveal the specific recognition motif of this enzyme.
XST1 and XST2: Tomato XyG Arabinofuranosyltransferases
In addition to the more common substituents found in XyG side chains, such as galactosyl and fucosyl residues, other identified sugar moieties include arabinopyranose (Selaginella kraussiana and Equisetum hyemale), galacturonic acid (Arabidopsis root hair and Physcomitrella patens; Peña et al., 2008, 2012), xylopyranose (Argania spinosa; Ray et al., 2004), and arabinofuranose (Ceratopteris richardii, tomato, and Olea europaea; Vierhuis et al., 2001; Jia et al., 2003; Peña et al., 2008). The differences in XyG structure among these plants are presumably caused by sequence differences in the GTs. The identification of such XyG GTs has been hampered by difficulties in generating or obtaining mutants in nonmodel species as well as a lack of genome sequence information. The recent completion of the tomato genome (Sato et al., 2012) allowed for the identification of candidate XyG arabinofuranosyltransferase genes. The overexpression of either XST1 or XST2 in the Arabidopsis mur3.1 xlt2 mutant background resulted in the production of arabinosylated XyG. While the activity of purified XST1 or XST2 was not demonstrated in vitro, the result that these proteins are sufficient for arabinosylation of XyG in planta and that the in vitro activity of the closely related AtMUR3 protein has been shown suggests that both XST1 and XST2 represent XyG arabinofuranosyltransferases. Interestingly, despite being more closely related to AtXLT2, which acts at the second position of the XXXG motif, both XST1 and XST2 resulted in the addition of arabinofuranose to the third position of the XXXG motif. The diversity of activities of various proteins within the MUR3 subclade of GT47, which can act at the first, second, or third position of the XXXG motif and can add Gal, galacturonic acid, arabinofuranose, and likely additional glycosyl groups, make it an interesting enzyme family to study for insight into the evolution of GT acceptor and donor specificities. Plant GTs believed to utilize UDP-arabinofuranose have previously been identified from the GT47 (Harholt et al., 2006), GT61 (Anders et al., 2012), and GT77 (Egelund et al., 2007; Gille et al., 2009) families, indicating that this donor specificity has evolved multiple times.
Origin of XST1 and XST2
A phylogenetic tree of approximately 400 angiosperm protein sequences from the GT47 family subclade including MUR3 and XLT2 (Li et al., 2004) suggests a specific subgroup of proteins (XST clade, which includes both XST1 and XST2; Fig. 2B) could represent XyG arabinofuranosyltransferases. This putative XST clade contains proteins from species within the Solanales, Lamiales, and Gentianales orders, all of which contain species with arabinosylated XyG (in Lamiales, O. europaea [Vierhuis et al., 2001] and in Gentianales, Nerium oleander [Hoffman et al., 2005]). While Gentianales and Lamiales species contain putative orthologs to AtXLT2, such a gene was not identified in any of the Solanales species. The absence of an AtXLT2 ortholog in the Solanales is consistent with the finding that none of the tomato genes expressed in the mur3.1 xlt2 mutant complemented the defect in XLT2 by resulting in the addition of a galactosyl residue to the middle xylosyl position of the XXXG motif. Together this suggests that a gene with XST activity arose following a gene duplication event in a common ancestor to the Lamiales, Gentianales, and Solanales and that the original XLT2 gene was subsequently lost in the Solanales lineage.
Implications for the Function of XyG Substitution
The mur3.1 xlt2 double mutant has approximately a 30% decrease in plant height at maturity and reduced cell wall creep. These phenotypes can be partially or fully rescued by expression of SlMUR3, XST1, or XST2. This demonstrates that for the function of XyG, the presence of a glycosyl substituent on the xylosyl residues is more important than the identity of that substituent. Previous studies showed that the enzymatic removal of side chains from XyG polysaccharides results in the formation of aggregates in vitro (Sims et al., 1998), demonstrating that XyG substitution can affect polymer solubility. XyG substitution may therefore help regulate the solubility of the XyG polysaccharide during biosynthesis and transport while still allowing for appropriate interactions with other polymers once deposited in the apoplast. Substitution is also a factor in XyG binding to cellulose and affects cellulose crystallinity (Lima et al., 2004; Whitney et al., 2006). Both of these processes affect cell wall structure and likely resulted in a wall with altered sensitivity to α-expansin-mediated creep, as seen in Figure 5C and also found previously in the xxt1/xxt2 line entirely lacking XyG (Park and Cosgrove, 2012a).
CONCLUSION
Identifying the genetic basis for and the functional significance of the structural diversity of plant cell wall polysaccharides remains a significant challenge due to the complex nature of their structures and the large number of putative carbohydrate active enzymes present in the typical plant genome, the majority of which are uncharacterized. When available, genetic approaches can be used to create mutants with reduced polysaccharide complexity. These mutants can then be used as platforms for the heterologous expression of GTs for identification and characterization as demonstrated here. Additionally, this approach allows for study of the in vivo function of particular cell wall structures.
Plant cell wall materials are important for a variety of applications including fuels, feed, food, textiles, and paper (Mishra and Malhotra, 2009), but native wall structures may not be ideal for these uses. Plant engineering approaches as presented here can allow for the tailoring of polysaccharide structures to produce glycans and wall materials with properties uniquely suited for particular applications. The naturally occurring diversity of wall components found in various plant species and tissues demonstrates the potential for altering wall structures for these purposes without detriment.
MATERIALS AND METHODS
Gene Identification and Phylogenetic Analysis
A BLAST (Altschul et al., 1997) search was performed using the Arabidopsis (Arabidopsis thaliana) protein sequences from the MUR3 and XLT2 subclade of GT family 47 to identify homologous genes from tomato (Solanum lycopersicum; Bombarely et al., 2011). Additional sequences were obtained from Phytozome (http://www.phytozome.net) and the Medicinal Plant Genomics Resource (http://medicinalplantgenomics.msu.edu/). A multiple sequence alignment was performed using Clustal (Sievers et al., 2011) and a maximum-likelihood tree was constructed using PhyML (Gouy et al., 2010; Guindon et al., 2010; Fig. 1).
Cloning and Transformation
The coding sequences of the GT47 candidate genes were amplified from genomic DNA using PCR with the primers from Supplemental Table S1 and cloned into the pCR8 TOPO TA vector with TA cloning (Life Technologies). LR reactions were performed to move the genes into a Gateway-compatible version of the plant overexpression plasmid pORE E4 (Coutu et al., 2007). These plasmids were transformed into Agrobacterium tumefaciens strain GV3101, which was subsequently used to transform the mur3.1 xlt2 Arabidopsis double mutant using the dip infiltration method (Clough and Bent, 1998). Transformed Arabidopsis plants were selected by plating the resulting seeds on one-half-strength Murashige and Skoog (Murashige and Skoog, 1962), 1% (w/v) Suc, and 60 µg mL–1 kanamycin agar plates. After 2 weeks, resistant plants were moved to soil and genotyped to confirm the presence of the transgene.
Xyloglucan Analysis
Analysis of XyG structure by XEG digestion of cell wall material followed by MALDI-TOF MS as previously described (Jensen et al., 2012).
Purification of the XXSG Oligosaccharide
To obtain large amounts of the initially unknown XyG oligosaccharide, 1-week-old etiolated hypocotyls were dried, ground, and extracted three times each with 70% (v/v) ethanol and chloroform:methanol (1:1, v/v). The pellet, approximately 150 mg, was then extracted with 4 m potassium hydroxide containing 10 mm sodium borohydride for 4 h under vigorous shaking. The supernatant was removed and neutralized with acetic acid and hydrochloric acid. Polysaccharides were precipitated from the neutralized extract by adding ethanol to 70% (v/v) and incubation of the solution at –20°C overnight. The precipitated material was washed four times with 70% (v/v) ethanol to remove remaining salts. The pellet was subsequently dried and digested with 4 mL of 50 mm ammonium formate, pH 4.5, containing 16 Units XEG. The supernatant from the digest was dried, and the oligosaccharides were reduced by incubation with 200 µL 1 m ammonium hydroxide containing 10 mg mL–1 sodium borohydride for 1 h. The reaction was neutralized with acetic acid followed by three additions and evaporations of methanol:acetic acid (9:1, v/v) and four additions of methanol. The reduced oligosaccharides were resuspended in 400 µL water and then separated using HPAEC-PAD. The fraction containing the dominant novel oligosaccharide (pooled from multiple injections) was desalted using a SupelClean ENVI-Carb 57109-U reversed-phase column (Supelco). The column was equilibrated with 50% (v/v) acetonitrile and washed four times with water. The oligosaccharide was bound to the column and then washed with water until the flow through showed neutral pH. The oligosaccharide was eluted from the column with 50% (v/v) acetonitrile and dried.
NMR
The purified oligosaccharide was dissolved in D2O (deuterium 99.99%), freeze dried, and dissolved in 0.3 mL D2O (deuterium 99.96%) with 3-(Trimethylsilyl)-1-propanesulfonic acid internal standard (0.01 mg mL–1). 1H-NMR spectra were recorded on a Bruker AVANCE 600 MHz NMR spectrometer equipped with an inverse gradient 5-mm TXI 1H/13C/15N CryoProbe at 285 K and 315 K, respectively. A two-dimensional Total Correlation Spectroscopy (TOCSY) NMR experiment was recorded using standard Bruker pulse program “dipsi2ph” for additional peak assignments with the following parameters: 2048 × 256 data points for F2 × F1 dimensions, 48 scans; four dummy scans; interscan delay of 1.5 s; prescan delay of 69.6 µs; and mixing time of 200 ms. All chemical shifts were referenced relative to 3-(Trimethylsilyl)-1-propanesulfonic acid (0.00 ppm for 1H). The NMR data processing and analysis were performed using Bruker’s Topspin 3.1 software.
Plant Growth Conditions and Phenotyping
The mur3.1 xlt2 mutant was generated as described in Jensen et al. (2012). Arabidopsis plants were grown on soil in a Percival growth chamber at 22°C with a repeating cycle of 16-h light and 8-h dark. Six-week-old plants were used for analysis of the growth phenotype. Plant height was measured from the base of the stem to the most distal part of the plant.
Wall Extension (Creep) Measurement
Eight-millimeter sections from the middle of petioles of the 4-week-old wild type and mutants were collected and prepared for an acid-induced wall extension (creep) assay and a stress strain assay as described (Park and Cosgrove, 2012a). For the creep assays, the clamped segments were initially incubated in neutral buffer (250 µL of 20 mm HEPES, pH 6.8) for 30 min with a constant load of 7.5 g. The incubation buffer was then replaced with 250 µL of 20 mm sodium acetate (pH 4.5) containing 5 mm dithiothreitol. Wall creep extension was measured with a position transducer, and the data were recorded every 30 s.
Sequence data referred to in this article can be found in the GenBank/EMBL data libraries under the following accession numbers: SlMUR3 (Sl09g064470; XP_004247129), XST1 (Sl07g044960; XP_004243615), XST2 (Sl07g049610; XP_004243499), Sl02g092840 (XP_004231889).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Quantification of transgene expression.
Supplemental Figure S2. XyG oligosaccharide mass profiles of independent transformants.
Supplemental Figure S3. HPAEC-PAD analysis of XEG-released XyG oligosaccharides.
Supplemental Figure S4. Glycosidic linkage analysis of the purified oligosaccharide.
Supplemental Figure S5. OREX digestion of the purified oligosaccharide.
Supplemental Figure S6. HPAEC-PAD analysis of XEG-released xyloglucan oligosaccharides from olive and the Arabidopsis mur3.1 xlt2 double mutant expressing XST1.
Supplemental Figure S7. 1H NMR peak assignments for the purified oligosaccharide.
Supplemental Figure S8. Representative pictures of Arabidopsis plants with altered xyloglucan structures.
Supplemental Figure S9. Extensibility of leaf petioles.
Supplemental Table S1. Primer sequences.
Acknowledgments
We thank Kirk Schnorr (Novozymes) for the generous gift of the XEG and Stefan Bauer (Energy Biosciences Institute, UC Berkeley) for a Pichia pastoris culture expressing the OREX enzyme.
Glossary
- XyG
xyloglucan
- XEG
xyloglucan-specific endoglucanase
- MALDI-TOF
matrix-assisted laser-desorption ionization-time-of-flight
- MS
mass spectrometry
- HPAEC
high-performance anion-exchange chromatography
- HPAEC-PAD
high-performance anion-exchange chromatography with pulsed amperometric detection
- m/z
mass-to-charge ratio
- GT
glycosyltransferase
References
- Aldington S, Mcdougall GJ, Fry SC. (1991) Structure-activity-relationships of biologically-active oligosaccharides. Plant Cell Environ 14: 625–636 [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders N, Wilkinson MD, Lovegrove A, Freeman J, Tryfona T, Pellny TK, Weimar T, Mortimer JC, Stott K, Baker JM, et al. (2012) Glycosyl transferases in family 61 mediate arabinofuranosyl transfer onto xylan in grasses. Proc Natl Acad Sci USA 109: 989–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer S, Vasu P, Mort AJ, Somerville CR. (2005) Cloning, expression, and characterization of an oligoxyloglucan reducing end-specific xyloglucanobiohydrolase from Aspergillus nidulans. Carbohydr Res 340: 2590–2597 [DOI] [PubMed] [Google Scholar]
- Bombarely A, Menda N, Tecle IY, Buels RM, Strickler S, Fischer-York T, Pujar A, Leto J, Gosselin J, Mueller LA. (2011) The Sol Genomics Network (solgenomics.net): growing tomatoes using Perl. Nucleic Acids Res 39: D1149–D1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier DM, Keegstra K. (2006) Two xyloglucan xylosyltransferases catalyze the addition of multiple xylosyl residues to cellohexaose. J Biol Chem 281: 34197–34207 [DOI] [PubMed] [Google Scholar]
- Cavalier DM, Lerouxel O, Neumetzler L, Yamauchi K, Reinecke A, Freshour G, Zabotina OA, Hahn MG, Burgert I, Pauly M, et al. (2008) Disrupting two Arabidopsis thaliana xylosyltransferase genes results in plants deficient in xyloglucan, a major primary cell wall component. Plant Cell 20: 1519–1537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cocuron JC, Lerouxel O, Drakakaki G, Alonso AP, Liepman AH, Keegstra K, Raikhel N, Wilkerson CG. (2007) A gene from the cellulose synthase-like C family encodes a β-1,4 glucan synthase. Proc Natl Acad Sci USA 104: 8550–8555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove DJ. (2000) Loosening of plant cell walls by expansins. Nature 407: 321–326 [DOI] [PubMed] [Google Scholar]
- Coutu C, Brandle J, Brown D, Brown K, Miki B, Simmonds J, Hegedus DD. (2007) pORE: a modular binary vector series suited for both monocot and dicot plant transformation. Transgenic Res 16: 771–781 [DOI] [PubMed] [Google Scholar]
- Egelund J, Obel N, Ulvskov P, Geshi N, Pauly M, Bacic A, Petersen BL. (2007) Molecular characterization of two Arabidopsis thaliana glycosyltransferase mutants, rra1 and rra2, which have a reduced residual arabinose content in a polymer tightly associated with the cellulosic wall residue. Plant Mol Biol 64: 439–451 [DOI] [PubMed] [Google Scholar]
- Faik A, Price NJ, Raikhel NV, Keegstra K. (2002) An Arabidopsis gene encoding an α-xylosyltransferase involved in xyloglucan biosynthesis. Proc Natl Acad Sci USA 99: 7797–7802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry SC, York WS, Albersheim P, Darvill A, Hayashi T, Joseleau JP, Kato Y, Lorences EP, Maclachlan GA, Mcneil M, et al. (1993) An unambiguous nomenclature for xyloglucan-derived oligosaccharides. Physiol Plant 89: 1–3 [Google Scholar]
- Gille S, de Souza A, Xiong G, Benz M, Cheng K, Schultink A, Reca IB, Pauly M. (2011) O-acetylation of Arabidopsis hemicellulose xyloglucan requires AXY4 or AXY4L, proteins with a TBL and DUF231 domain. Plant Cell 23: 4041–4053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gille S, Hänsel U, Ziemann M, Pauly M. (2009) Identification of plant cell wall mutants by means of a forward chemical genetic approach using hydrolases. Proc Natl Acad Sci USA 106: 14699–14704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouy M, Guindon S, Gascuel O. (2010) SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol 27: 221–224 [DOI] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59: 307–321 [DOI] [PubMed] [Google Scholar]
- Hantus S, Pauly M, Darvill AG, Albersheim P, York WS. (1997) Structural characterization of novel l-galactose-containing oligosaccharide subunits of jojoba seed xyloglucans. Carbohydr Res 304: 11–20 [DOI] [PubMed] [Google Scholar]
- Harholt J, Jensen JK, Sørensen SO, Orfila C, Pauly M, Scheller HV. (2006) ARABINAN DEFICIENT 1 is a putative arabinosyltransferase involved in biosynthesis of pectic arabinan in Arabidopsis. Plant Physiol 140: 49–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T, Kaida R. (2011) Functions of xyloglucan in plant cells. Mol Plant 4: 17–24 [DOI] [PubMed] [Google Scholar]
- Hoffman M, Jia Z, Peña MJ, Cash M, Harper A, Blackburn AR, II, Darvill A, York WS. (2005) Structural analysis of xyloglucans in the primary cell walls of plants in the subclass Asteridae. Carbohydr Res 340: 1826–1840 [DOI] [PubMed] [Google Scholar]
- Jensen JK, Schultink A, Keegstra K, Wilkerson CG, Pauly M. (2012) RNA-Seq analysis of developing nasturtium seeds (Tropaeolum majus): identification and characterization of an additional galactosyltransferase involved in xyloglucan biosynthesis. Mol Plant 5: 984–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Z, Cash M, Darvill AG, York WS. (2005) NMR characterization of endogenously O-acetylated oligosaccharides isolated from tomato (Lycopersicon esculentum) xyloglucan. Carbohydr Res 340: 1818–1825 [DOI] [PubMed] [Google Scholar]
- Jia Z, Qin Q, Darvill AG, York WS. (2003) Structure of the xyloglucan produced by suspension-cultured tomato cells. Carbohydr Res 338: 1197–1208 [DOI] [PubMed] [Google Scholar]
- Lerouxel O, Choo TS, Séveno M, Usadel B, Faye L, Lerouge P, Pauly M. (2002) Rapid structural phenotyping of plant cell wall mutants by enzymatic oligosaccharide fingerprinting. Plant Physiol 130: 1754–1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Cordero I, Caplan J, Mølhøj M, Reiter WD. (2004) Molecular analysis of 10 coding regions from Arabidopsis that are homologous to the MUR3 xyloglucan galactosyltransferase. Plant Physiol 134: 940–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima DU, Loh W, Buckeridge MS. (2004) Xyloglucan-cellulose interaction depends on the sidechains and molecular weight of xyloglucan. Plant Physiol Biochem 42: 389–394 [DOI] [PubMed] [Google Scholar]
- Lopes FJF, Pauly M, Brommonshenkel SH, Lau EY, Diola V, Passos JL, Loureiro ME. (2010) The EgMUR3 xyloglucan galactosyltransferase from Eucalyptus grandis complements the mur3 cell wall phenotype in Arabidopsis thaliana. Tree Genet Genomes 6: 745–756 [Google Scholar]
- Madson M, Dunand C, Li X, Verma R, Vanzin GF, Caplan J, Shoue DA, Carpita NC, Reiter WD. (2003) The MUR3 gene of Arabidopsis encodes a xyloglucan galactosyltransferase that is evolutionarily related to animal exostosins. Plant Cell 15: 1662–1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra A, Malhotra AV. (2009) Tamarind xyloglucan: a polysaccharide with versatile application potential. J Mater Chem 19: 8528–8536 [Google Scholar]
- Murashige T, Skoog F. (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Park YB, Cosgrove DJ. (2012a) Changes in cell wall biomechanical properties in the xyloglucan-deficient xxt1/xxt2 mutant of Arabidopsis. Plant Physiol 158: 465–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park YB, Cosgrove DJ. (2012b) A revised architecture of primary cell walls based on biomechanical changes induced by substrate-specific endoglucanases. Plant Physiol 158: 1933–1943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauly M, Andersen LN, Kauppinen S, Kofod LV, York WS, Albersheim P, Darvill A. (1999) A xyloglucan-specific endo-β-1,4-glucanase from Aspergillus aculeatus: expression cloning in yeast, purification and characterization of the recombinant enzyme. Glycobiology 9: 93–100 [DOI] [PubMed] [Google Scholar]
- Pauly M, Gille S, Liu L, Mansoori N, de Souza A, Schultink A, Xiong G. (June 26, 2013) Hemicellulose biosynthesis. Planta [DOI] [PubMed] [Google Scholar]
- Pauly M, Keegstra K. (2008) Cell-wall carbohydrates and their modification as a resource for biofuels. Plant J 54: 559–568 [DOI] [PubMed] [Google Scholar]
- Peña MJ, Darvill AG, Eberhard S, York WS, O’Neill MA. (2008) Moss and liverwort xyloglucans contain galacturonic acid and are structurally distinct from the xyloglucans synthesized by hornworts and vascular plants. Glycobiology 18: 891–904 [DOI] [PubMed] [Google Scholar]
- Peña MJ, Kong Y, York WS, O’Neill MA. (2012) A galacturonic acid-containing xyloglucan is involved in Arabidopsis root hair tip growth. Plant Cell 24: 4511–4524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrin RM, DeRocher AE, Bar-Peled M, Zeng WQ, Norambuena L, Orellana A, Raikhel NV, Keegstra K. (1999) Xyloglucan fucosyltransferase, an enzyme involved in plant cell wall biosynthesis. Science 284: 1976–1979 [DOI] [PubMed] [Google Scholar]
- Ray B, Loutelier-Bourhis C, Lange C, Condamine E, Driouich A, Lerouge P. (2004) Structural investigation of hemicellulosic polysaccharides from Argania spinosa: characterisation of a novel xyloglucan motif. Carbohydr Res 339: 201–208 [DOI] [PubMed] [Google Scholar]
- Sampedro J, Cosgrove DJ. (2005) The expansin superfamily. Genome Biol 6: 242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato S, Tabata S, Hirakawa H, Asamizu E, Shirasawa K, Isobe S, Kaneko T, Nakamura Y, et al. (2012) The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485: 635–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheller HV, Ulvskov P. (2010) Hemicelluloses. Annu Rev Plant Biol 61: 263–289 [DOI] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, et al. (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7: 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims IM, Gane AM, Dunstan D, Allan GC, Boger DV, Melton LD, Bacic A. (1998) Rheological properties of xyloglucans from different plant species. Carbohydr Polym 37: 61–69 [Google Scholar]
- Somerville C, Bauer S, Brininstool G, Facette M, Hamann T, Milne J, Osborne E, Paredez A, Persson S, Raab T, et al. (2004) Toward a systems approach to understanding plant cell walls. Science 306: 2206–2211 [DOI] [PubMed] [Google Scholar]
- Tamura K, Shimada T, Kondo M, Nishimura M, Hara-Nishimura I. (2005) KATAMARI1/MURUS3 Is a novel golgi membrane protein that is required for endomembrane organization in Arabidopsis. Plant Cell 17: 1764–1776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanzin GF, Madson M, Carpita NC, Raikhel NV, Keegstra K, Reiter WD. (2002) The mur2 mutant of Arabidopsis thaliana lacks fucosylated xyloglucan because of a lesion in fucosyltransferase AtFUT1. Proc Natl Acad Sci USA 99: 3340–3345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierhuis E, York WS, Kolli VS, Vincken J, Schols HA, Van Alebeek GW, Voragen AG. (2001) Structural analyses of two arabinose containing oligosaccharides derived from olive fruit xyloglucan: XXSG and XLSG. Carbohydr Res 332: 285–297 [DOI] [PubMed] [Google Scholar]
- Whitney SEC, Wilson E, Webster J, Bacic A, Reid JSG, Gidley MJ. (2006) Effects of structural variation in xyloglucan polymers on interactions with bacterial cellulose. Am J Bot 93: 1402–1414 [DOI] [PubMed] [Google Scholar]
- Zabotina OA. (2012) Xyloglucan and its biosynthesis. Front Plant Sci 3: 134. [DOI] [PMC free article] [PubMed] [Google Scholar]