Suppression of either of two wheat genes decreases the amount of arabinoxylan, the major cell wall polymer in wheat flour, by 50%.
Abstract
The cell walls of wheat (Triticum aestivum) starchy endosperm are dominated by arabinoxylan (AX), accounting for 65% to 70% of the polysaccharide content. Genes within two glycosyl transferase (GT) families, GT43 (IRREGULAR XYLEM9 [IRX9] and IRX14) and GT47 (IRX10), have previously been shown to be involved in the synthesis of the xylan backbone in Arabidopsis, and close homologs of these have been implicated in the synthesis of xylan in other species. Here, homologs of IRX10 TaGT47_2 and IRX9 TaGT43_2, which are highly expressed in wheat starchy endosperm cells, were suppressed by RNA interference (RNAi) constructs driven by a starchy endosperm-specific promoter. The total amount of AX was decreased by 40% to 50% and the degree of arabinosylation was increased by 25% to 30% in transgenic lines carrying either of the transgenes. The cell walls of starchy endosperm in sections of grain from TaGT43_2 and TaGT47_2 RNAi transgenics showed decreased immunolabeling for xylan and arabinoxylan epitopes and approximately 50% decreased cell wall thickness compared with controls. The proportion of AX that was water soluble was not significantly affected, but average AX polymer chain length was decreased in both TaGT43_2 and TaGT47_2 RNAi transgenics. However, the long AX chains seen in controls were absent in TaGT43_2 RNAi transgenics but still present in TaGT47_2 RNAi transgenics. The results support an emerging picture of IRX9-like and IRX10-like proteins acting as key components in the xylan synthesis machinery in both dicots and grasses. Since AX is the main component of dietary fiber in wheat foods, the TaGT43_2 and TaGT47_2 genes are of major importance to human nutrition.
Xylan is a hemicellulosic component of cell walls and one of the most abundant polysaccharides in nature (Ebringerova et al., 2005; Scheller and Ulvskov, 2010). Its prevalence and structure differ markedly between dicots and grasses; in the former, it constitutes only about 5% of the polysaccharide of primary cell walls, whereas it is typically 30% of the polysaccharide of grass primary cell walls (Carpita, 1996; Scheller and Ulvskov, 2010). Arabinofuranose (Araf) substitution of the xylan backbone is more common in grasses than in dicots, and some of the Araf sugars are ester linked to ferulic acid, which confers a cross-linking functionality to the xylan chains that is absent in dicots. Furthermore, the reducing end of dicot xylan has a characteristic 4-β-d-Xylp-(1→4)-β-d-Xylp-(1→3)-α-l-Rhap(1→2)-α-d-GalpA-(1→4)-d-Xylp oligosaccharide that has not been detected in xylans from grasses (Pena et al., 2007; York and O’Neill, 2008).
The candidate genes responsible for xylan synthesis in Arabidopsis (Arabidopsis thaliana) have been identified by studies of knockout mutants in glycosyl transferase (GT) families. Plants carrying mutations in the GT43 family genes IRREGULAR XYLEM9 (IRX9) and IRX14 and in the GT47 family gene IRX10 all have decreased xylan content, with the remaining xylan having a shorter chain length compared with the wild type but the reducing end oligosaccharide still being present (Brown et al., 2007, 2009; Pena et al., 2007; Wu et al., 2009). Closely related genes exist for all these (IRX9-L, IRX10-L, and IRX14-L) but are less expressed and are functionally redundant with the more highly expressed counterpart (Brown et al., 2009; Wu et al., 2010). It is unclear how the products from the very different GT43 and GT47 gene families, which appear to be equally essential in Arabidopsis, cooperate to synthesize the xylan backbone. By overexpressing IRX9 and IRX14 together in tobacco (Nicotiana tabacum) BY2 cells, xylan xylosyl transferase activity was stimulated, suggesting that they encode the key proteins mediating the activity, although an endogenous IRX10 ortholog would presumably also be present in these cells (Lee et al., 2012). On the other hand, abundant xylan is synthesized in psyllium (Plantago afra) seed mucilage, which has abundant transcripts of an IRX10 ortholog but only very low levels of transcripts of IRX9 and IRX14 orthologs (Jensen et al., 2011). It is possible that different xylan synthetic mechanisms exist, for example between grasses and dicots (York and O’Neill, 2008).
There is less direct evidence for the genes involved in xylan backbone synthesis in grasses. Recently, a rice (Oryza sativa) line carrying a mutation in the ortholog of IRX10 was shown to have decreased xylan content, although the decrease was modest (10% decrease in cell wall Xyl). The mutation also resulted in smaller stature and increased ease of biomass saccharification (Chen et al., 2013). A microsomal complex isolated from wheat (Triticum aestivum) seedlings that exhibited all the main activities required for synthesizing glucuronoarabinoxylan (i.e. xylan backbone extension and addition of Araf and GlcA sugars to the backbone) was shown to contain three wheat proteins that were homologous to the GT43 IRX14, the GT47 IRX10, and a GT75 protein (Zeng et al., 2010). This GT75 protein is orthologous to OsUAM1 and presumably has the same UDP-Ara mutase activity found for the protein in rice (Konishi et al., 2007), converting the supplied UDP-Arap into the UDP-Araf found in the products. Since the observed xylan GlcA transferase activity is retaining and the three identified proteins are all from inverting families, other active proteins must be present. In Arabidopsis, xylan GlcA transferase proteins from GT8 have been shown to confer this activity (Mortimer et al., 2010). Also, GT61 proteins have been shown to be xylan arabinosyl transferases in grasses, including wheat (Anders et al., 2012). While only a IRX14 homolog was detected in this complex, of great interest is whether a IRX9 homolog is also required in grasses for xylan backbone extension, as appears to be the case in woody and herbaceous dicots (Brown et al., 2007; Pena et al., 2007; Lee et al., 2011, 2012).
The wheat starchy endosperm cell wall has been established as an excellent system for studying grass cell walls due to the prevalence of arabinoxylan (AX) and 1,3;1,4-β-d-glucan, which constitute approximately 70% and 25% of the total cell wall polysaccharide, respectively. Using a strong starchy endosperm-specific promoter to drive RNA interference (RNAi) to suppress specific transcripts, we have demonstrated the key role of TaCSLF6 in synthesizing 1,3;1,4-β-d-glucan (Nemeth et al., 2010) and that the GT61 gene TaXAT1 is responsible for nearly all the monosubstituted Ara in AX in endosperm (Anders et al., 2012). The amount and structure of AX in wheat starchy endosperm are also of great practical importance; as it is the main component of dietary fiber in wheat foods, it is a major contributor of dietary fiber in the human diet (Topping, 2007), and it also affects the processing properties of wheat flour for different end uses (Saulnier et al., 2007).
The most abundant transcripts for the genes implicated in xylan backbone extension (xylan synthases) in wheat starchy endosperm are TaGT47_2, TaGT43_1, and TaGT43_2, which are homologous to IRX10, IRX14, and IRX9, respectively, in Arabidopsis (Pellny et al., 2012). In fact, GT47_2 is the most abundantly expressed of all GT genes in this cell type (excluding the genes involved in starch synthesis), in keeping with the dominance of AX in the cell wall. Here, we report the effects of specifically suppressing the expression of TaGT47_2 and TaGT43_2 by RNAi in transgenic wheat lines on the amount and structure of AX and the wheat starchy endosperm cell wall. The effects of the suppression are similar for these two diverse genes, suggesting an analogous mechanism for xylan extension in wheat to that in Arabidopsis.
RESULTS
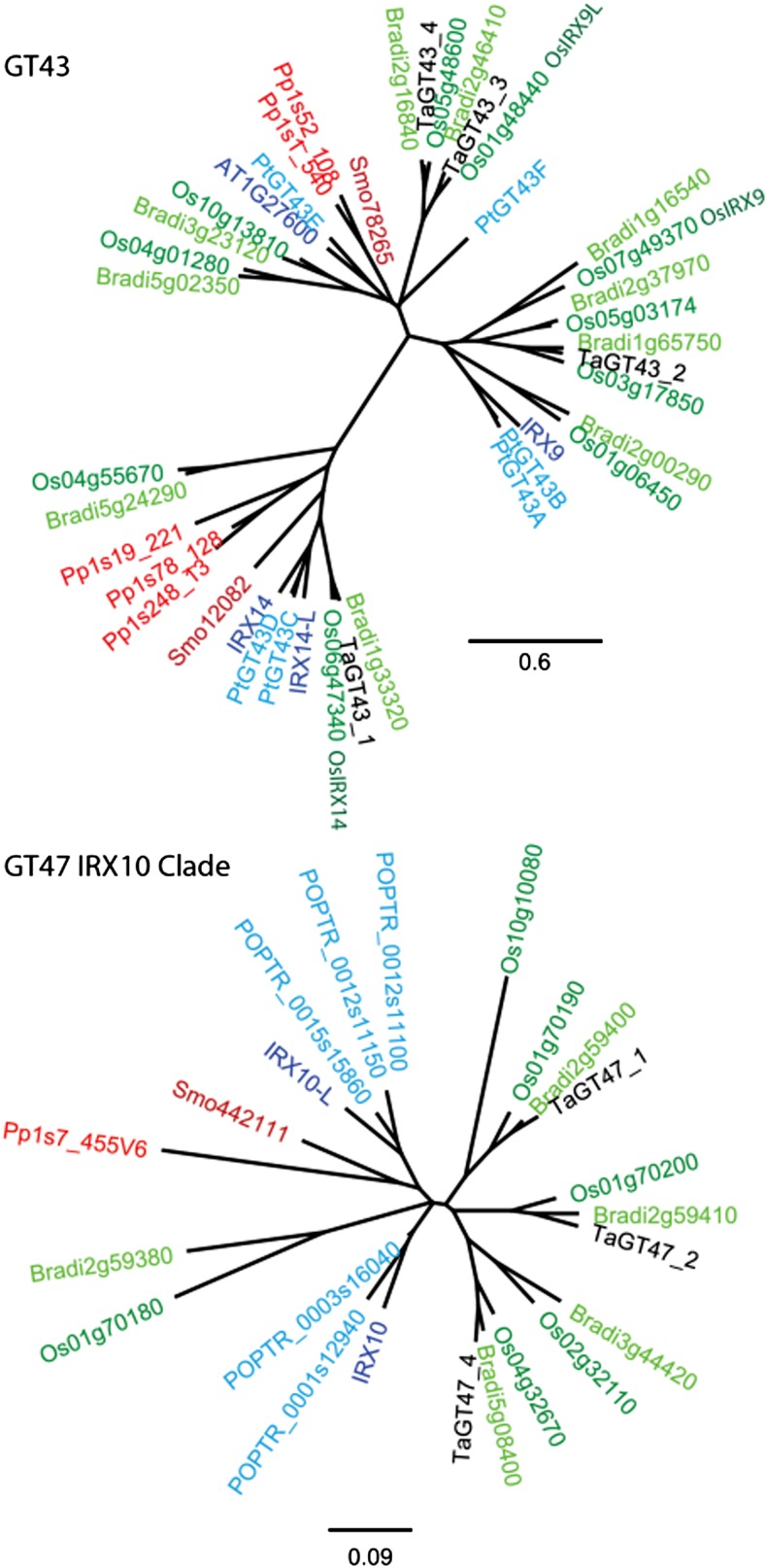
Phylogenetic trees for predicted protein sequences for the whole GT43 family and for the IRX10 clade of the GT47 family from the fully sequenced genomes of Arabidopsis, poplar (Populus trichocarpa), rice, Brachypodium distachyon, Physcomitrella patens, and Selaginella moellendorffii (www.phytozome.org; Goodstein et al., 2012) and from wheat transcripts present in the starchy endosperm (Pellny et al., 2012) are shown in Figure 1. Three rice genes in the GT43 family are indicated that have recently been demonstrated to be functional orthologs of IRX9, IRX9-L, and IRX14, being able to complement mutations in these Arabidopsis genes (Chiniquy et al., 2013). While there is considerable diversity between the IRX9 and IRX14 homologs, the IRX10 homologs are highly conserved (note the different scale bars for the two trees). The two wheat genes studied here are the IRX9 homolog TaGT43_2 and the IRX10 homolog TaGT47_2.
Figure 1.
Phylogenetic trees of the GT43 family and the IRX10 clade within the GT47 family. Trees were derived from protein alignments of all genes of fully sequenced plants plus wheat genes expressed in starchy endosperm. Lower plants are represented by S. moellendorffii (dark red) and P. patens (orange); monocots by rice (dark green), B. distachyon (light green), and wheat genes (black); and dicots by Arabidopsis (dark blue) and poplar (light blue).
Bread wheat is a hexaploid with three related genomes (A, B, and D). The term “gene,” therefore, is used to encompass the three homeologous forms from the three genomes; these typically have 95% to 97% nucleotide identity of transcripts and can be assumed to have the same molecular function. The recent availability of the chromosome-sorted genomic survey sequences from the International Wheat Genome Sequencing Consortium (www.wheatgenome.org) makes it possible to unequivocally assign the three variants in complementary DNA sequence that we identify for each of the two genes to chromosomes. Therefore, TaGT43_2 and TaGT47_2 genes can be assigned to chromosomes 4 and 3 (i.e. 4A, 4B, 4D and 3A, 3B, 3D), respectively, with all three homeologs of both being expressed in starchy endosperm. The transcript abundances differ somewhat between the three homeologs but all show the same pattern through endosperm development for both genes (Fig. 2). These sequences and the RNAi constructs designed to suppress all three variants are shown in Supplemental Figures S1 and S2. The RNAi construct for TaGT47_2 had sufficient identity to potentially also suppress the very similar TaGT47_1 and TaGT47_4 genes (whose longest identical fragments are 29 and 38 bp, respectively), whereas the RNAi construct for TaGT43_2 was specific for this gene.
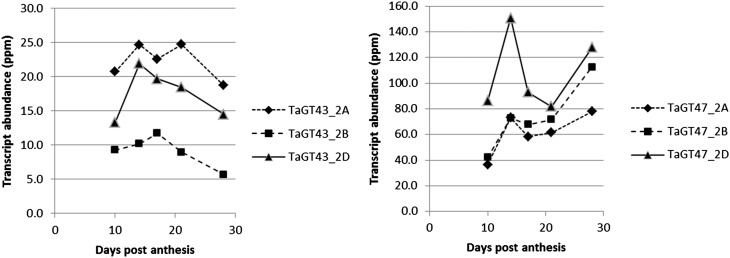
Figure 2.
Transcript abundance of TaGT43_2 and TaGT47_2 in developing starchy endosperm. Estimates were based on reanalysis of transcript abundance for three homeologs of TaGT43_2 and TaGT47_2 from RNA-Seq libraries (Pellny et al., 2012).
We identified six wheat transgenic lines carrying the TaGT43_2 RNAi transgene and six lines carrying the TaGT47_2 RNAi. All the TaGT43_2 RNAi lines had clear effects on the amounts of arabinoxylan oligosaccharide (AXOS) released by xylanase digestion in samples from T1 grain, but three of the GT47_2 RNAi lines did not show any clear effects on AXOS in T1 samples and were not pursued further (data not shown). Such an absence of effect is sometimes encountered in cereal transformants due to the partial integration of transgenes or integration into a nonexpressed part of the genome. Data are presented for four GT43_2 RNAi lines and the three GT47_2 RNAi lines that had an effect; all these exhibited a segregation of transgenes consistent with a single insertion site. For all measurements, samples for homozygous transgenic plants were compared with corresponding azygous null segregants from the same line.
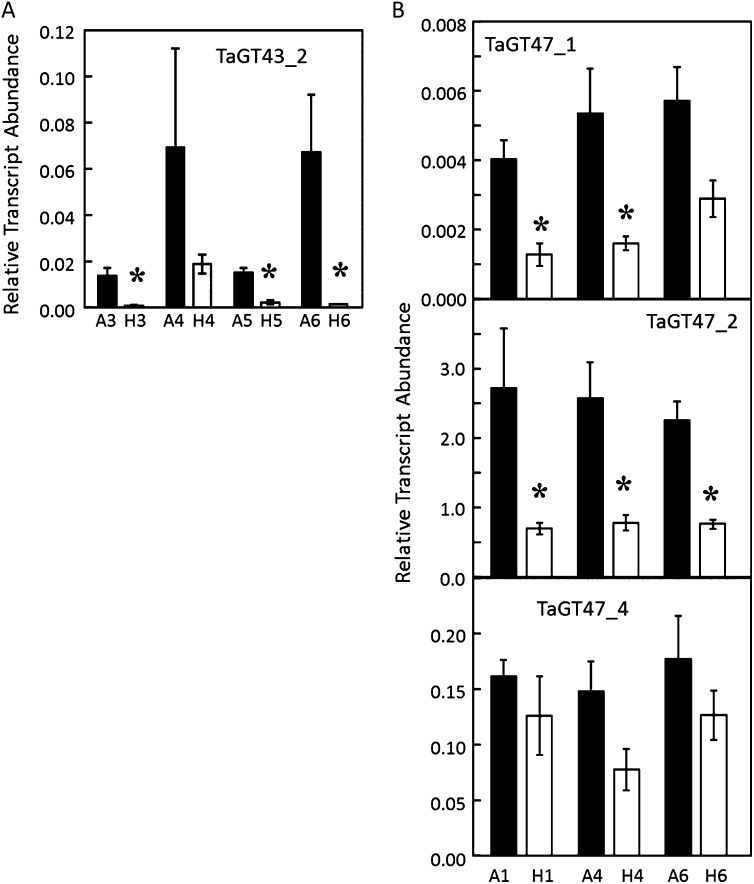
Transcript abundance within the developing starchy endosperm of the transgenic lines was determined by quantitative reverse transcription-PCR, showing strong suppression of the target genes in all four GT43_2 RNAi lines and the three GT47_2 RNAi lines (Fig. 3). However, there was also suppression of the closely related GT47_1 gene in the GT47_2 RNAi lines and some evidence of GT47_4 suppression. The phenotypes presented for these GT47_2 RNAi lines, therefore, may partially result from the suppression of GT47_1 and GT47_4, although these genes are expressed at only approximately 10% of the level of the GT47_2 genes in starchy endosperm (Pellny et al., 2012). These three very similar genes appear to have arisen from duplication after divergence from the common ancestor with IRX10, and it is not possible to say which of them is the true IRX10 ortholog (Fig. 1).
Figure 3.
Effects of RNAi transgenes on endogenous transcript abundance. Transcript abundance was estimated by quantitative reverse transcription-PCR from pure starchy endosperm dissected at 21 DPA. Error bars represent se, and stars denote significant differences at P < 0.05. A, Four replicate samples from homozygous plants carrying the TaGT43_2 RNAi transgene for lines 3, 4, 5, and 6 (H3, H4, H5, and H6) or the corresponding azygous controls (A3, A4, A5, and A6). B, As in A, but for the TaGT47_2 RNAi lines 1, 4, and 6.
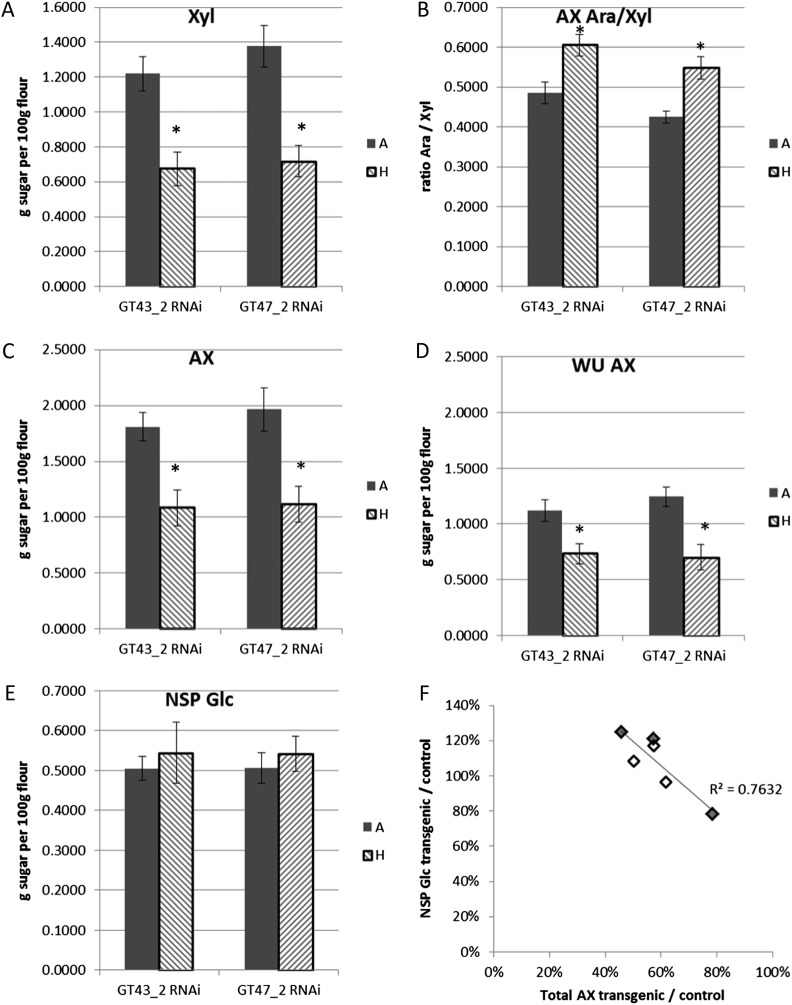
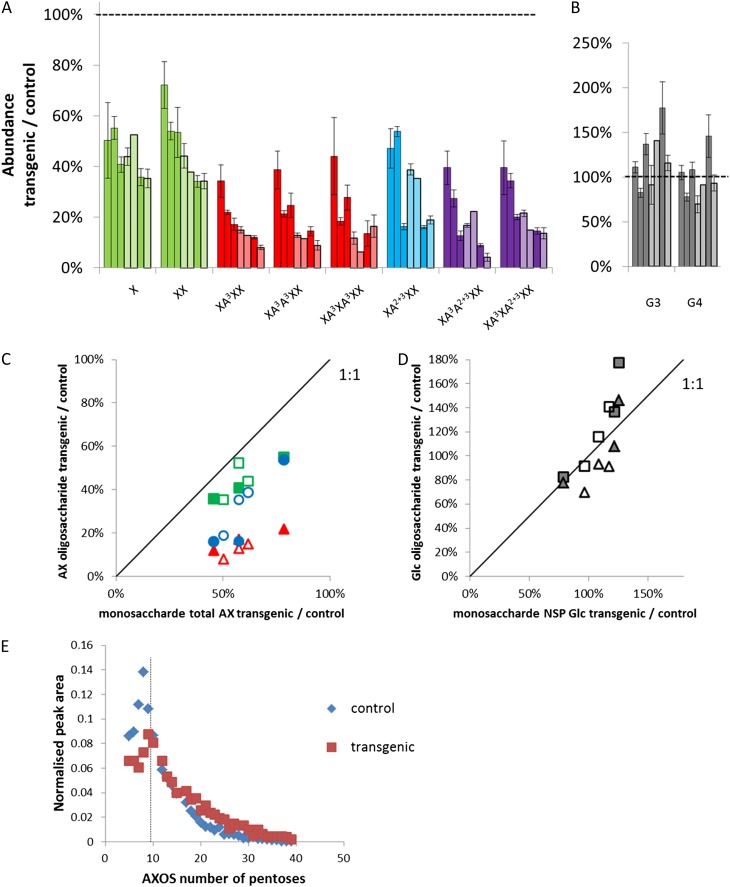
Monosaccharide analyses of nonstarch polysaccharides (NSP; composed of cell wall polysaccharides, oligosaccharides, and arabinogalactan peptide) from white flour showed that suppression of TaGT43_2 and TaGT47_2 decreased the total amount of NSP Xyl, which is virtually all in the AX backbone (Ordaz-Ortiz and Saulnier, 2005), by 45% and 48%, respectively, averaged across three lines (Fig. 4A). There was an increase in the ratio of AX Ara to Xyl in the transgenic lines, indicating greater Ara substitution of the remaining Xyl residues (Fig. 4B). Thus, total AX was decreased by 40% and 43% (Fig. 4C). Similar effects were observed for the water-unextractable portion of AX (Fig. 4D), showing that there was no overall effect on the solubility of AX, with water-extractable (WE) AX being about 35% of total AX in all lines. There was a small increase in the amount of Glc in the NSP in all of the transgenic lines relative to the controls (Fig. 4E). Although this was not significant when averaged across the three lines for each gene, there was a clear trend, with the lines showing the greatest decrease in AX also having the greatest increase in NSP Glc (Fig. 4F). However, this only had a minor influence on the total amount of cell wall polysaccharide, the lines with a decrease of approximately 50% AX having an increase in Glc of approximately 20%, which would give an overall decrease in cell wall sugars of approximately 30%. There was no significant trend in the amount of Man that was present in the NSP, suggesting that glucomannan was not affected (Supplemental Table S1).
Figure 4.
Monosaccharide analyses of NSP fractions from white flour. All data are from samples from three TaGT43_2 RNAi lines (lines 1, 3, and 6) and three TaGT47_2 RNAi lines (lines 1, 4, and 6). A to E, Average of azygous controls (A) and homozygous transgenics (H) for three lines; error bars represent se, and asterisks denote significant differences at P < 0.05 from Student’s t test. A, Total Xyl, assumed to be all in AX. B, Ratio of Ara to Xyl in AX. Ara in AX estimated as (Ara \x{2013} 0.7 × Gal), since it is assumed all Gal is in AGP, with AGP Ara/Gal = 0.7 (Ordaz-Ortiz and Saulnier, 2005). C, Total AX, estimated as Xyl + Ara in AX, estimated as in B. D, Water-unextractable (WU) AX, estimated as for total AX, using monosaccharide values from the water-unextractable fraction. E, Total Glc. F, Effect of transgenes on total AX versus effect on NSP Glc, expressed as values for transgenics relative to corresponding controls in individual lines. Gray symbols represent TaGT43_2 lines, and white symbols represent TaGT47_2 lines.
AX structure was characterized using simultaneous digestion of NSP preparations of white flour by xylanase and lichenase and analysis of the resulting oligosaccharides as described previously (Ordaz-Ortiz et al., 2005; Nemeth et al., 2010; Pellny et al., 2012). Large decreases (significant at P < 0.001) were observed in the abundances of AXOS in all of the transgenic lines (Fig. 5A). However, the magnitude of effects differed between individual fragments, with unsubstituted Xyl and xylobiose (X and XX) being affected less than substituted AXOS, and XA2+3XX being affected slightly less than other substituted AXOS. The effects on glucan oligosaccharides (G3 and G4) released by lichenase digestion were more variable: in some lines, significant increases were observed, while in others, no effect or small decreases were seen (Fig. 5B); these variable effects were much the same as those for NSP Glc (Fig. 5D), indicating that the effects on Glc are largely or solely due to changes in the 1,3;1,4-β-d-glucan from which the G3 and G4 oligosaccharides are derived. The magnitude of the effects on AXOS across the lines was correlated with the size of the effect on total AX estimated from monosaccharide analysis, with correlation coefficients of 0.83, 0.87, and 0.90 for X, XA3XX, and XA2+3XX, respectively. However, for all lines, the relative decrease in AXOS peaks was greater than the decrease in total AX measured as a monosaccharide (e.g. a line with approximately 50% decrease in total AX had a decrease of approximately 90% in XA3XX; Fig. 5D). Virtually all the water-unextractable portion of AX was solubilized by xylanase action, and this was unchanged in the transgenics (Supplemental Table S1). However, our high-performance anion-exchange chromatography (HPAEC) methodology measures the abundance of AXOS containing up to nine pentoses, accounting for about 75% of the total in wild-type wheat, but matrix-assisted laser-desorption ionization (MALDI)-mass spectrometry (MS) analysis of digested AX shows that in transgenic samples, this fraction was much smaller (Fig. 5E), explaining the greater decrease in measured AXOS. This is presumably caused by the greater arabinosylation of AX in transgenic samples (Fig. 4B), which results in fewer consecutive unsubstituted Xyl residues that are required for GH11 xylanase cleavage.
Figure 5.
Relative abundance of oligosaccharides released by simultaneous digestion of endosperm NSP samples by xylanase and lichenase. A to D, Oligosaccharide abundance from HPAEC peak area for transgenics relative to corresponding controls in individual lines. A and B, Relative abundance (average of three biological replicates ± se). Solid colors represent GT43_2 lines, and striped colors represent GT47_2 lines. Columns are sorted by decreasing relative abundance of XA3XX: GT43_2 lines 2, 6, and 1, GT47_2 lines 1 and 6, GT43_2 line 3, and GT47_2 line 4. A, AXOS. Nomenclature is from Faure et al. (2009). Columns are colored according to substitution with Araf: unsubstituted (green), monosubstituted only (red), disubstituted only (blue), and monosubstituted and disubstituted (purple). B, (1,3);(1,4)-β-Glucan oligosaccharides [G3, (Glc)3; G4, (Glc)4]. C and D, Comparison of relative oligosaccharide and monosaccharide abundances for individual lines. Colored symbols represent GT43_2 lines, and white symbols represent GT47_2 lines. C, Relative total AX abundance from monosaccharide analyses versus relative AXOS abundance released by xylanase. Squares represent X, triangles represent XA3XX, and circles represent XA2+3XX; colors are as in A. D, Relative NSP Glc abundance versus relative (1,3);(1,4)-β-glucan oligosaccharide abundance. Squares represent G3, and triangles represent G4. E, Relative distribution of AXOS released by xylanase estimated by MALDI-MS. Number of pentoses (DP) was identified from mass-to-charge ratio = DP × 132.114 + 18.016 + 23. Data are from transgenic and corresponding null samples of GT43_2 line 3. Peaks at DP 11, 16, and 27 are omitted because of overlap with hexose oligosaccharides; AXOS of DP < 5 was not analyzed. The dotted line indicates DP below which AXOS was analyzed by the HPAEC method.
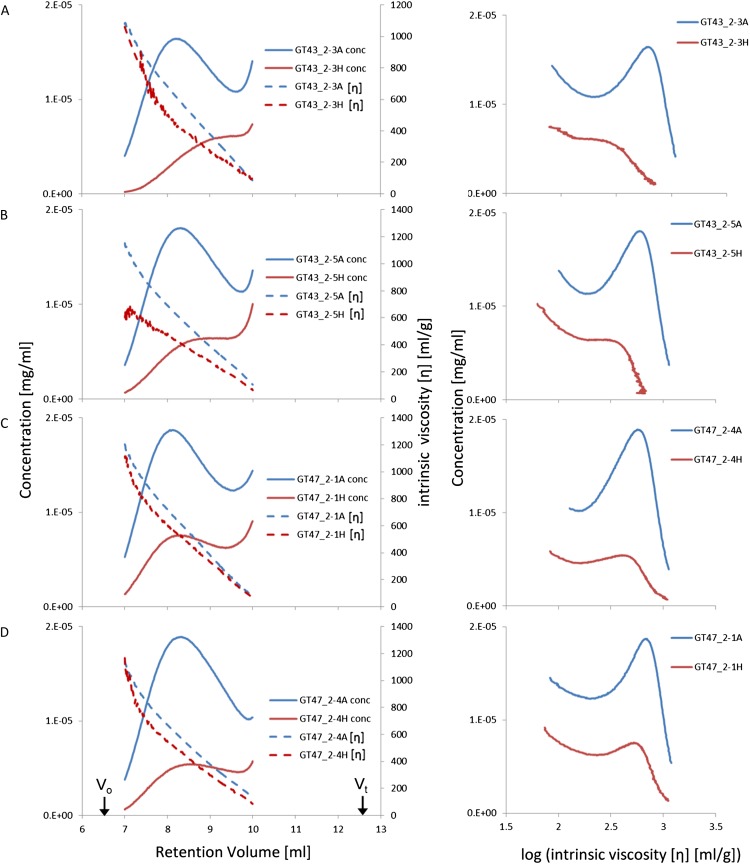
Since Arabidopsis mutants lacking functional IRX9 or IRX10 have decreased glucuronoxylan chain length (Brown et al., 2007, 2009; Pena et al., 2007), we determined the distribution of WE-AX chain size by measuring the concentration and intrinsic viscosity ([η]) profiles of WE-AX separated by size-exclusion HPLC. The concentration profiles for samples from two homozygous transgenic lines each of GT43_2 RNAi (lines 3 and 5) and GT47_2 RNAi (lines 1 and 4) showed shifts to higher retention volumes compared with azygous controls, indicating that, in all cases, a greater fraction of the WE-AX was made up of small molecules (Fig. 6). This reflects a shift in Mr distribution, which depends both on chain length and Ara substitution, but [η] shows a close direct relationship with AX chain length, as log [η] is linearly related to log chain length (Dervilly-Pinel et al., 2001). The profiles of the transgenic samples show much greater relative abundance of AX at low [η], consistent with a shift to shorter AX chains (Fig. 6) and resulting in a pronounced decrease in average [η] (Table I). However, comparison of the log [η] value in the control and transgenic plants shows different effects between the GT43_2 RNAi and GT47_2 RNAi constructs. In particular, the peak in log [η] of about 2.8 to 2.9 in the controls is shifted to a shoulder at 2.5 to 2.6 in the GT43_2 RNAi transgenics (Fig. 6, A and B) but not in the GT47_2 RNAi transgenics (Fig. 6, C and D). Based on the relationship between log [η] and log chain length reported by Dervilly-Pinel et al. (2001), the effect of the GT43_2 RNAi construct corresponds to a decrease in AX chain length at this peak/shoulder from 1,900 to 2,300 down to 750 to 950 Xyl residues.
Figure 6.
Profiles of concentration and [η] from size-exclusion chromatography for WE-AX from mature endosperm of transgenic and control wheat lines. Left panels show original profiles against retention volume, with void volume (Vo) and total volume (Vt) of the column indicated. Right panels show log [η], which is linearly related to log AX chain length, versus concentration. Data are shown for TaGT43_2 RNAi lines 3 and 5 and for TaGT47_2 RNAi lines 1 and 4 azygous control (A) and homozygous transgenic (H) samples.
Table I. [η] and amount (percentage of white flour) of WE-AX from homozygous and azygous wheat lines determined by HPSEC.
The values are averages of the profiles against retention volume shown in Figure 6. Results are presented for each of two extractions (1 and 2).
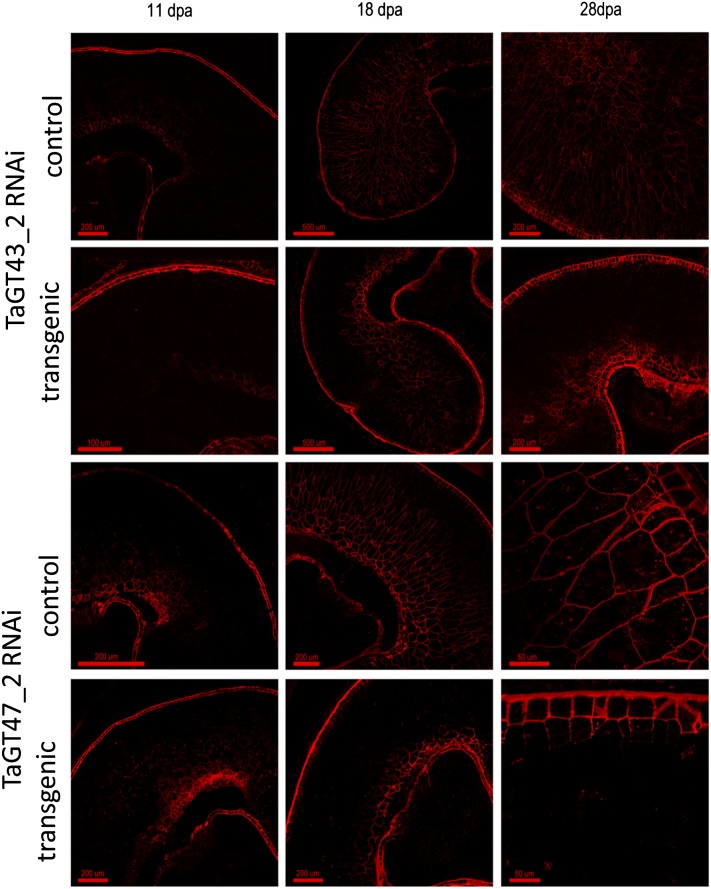
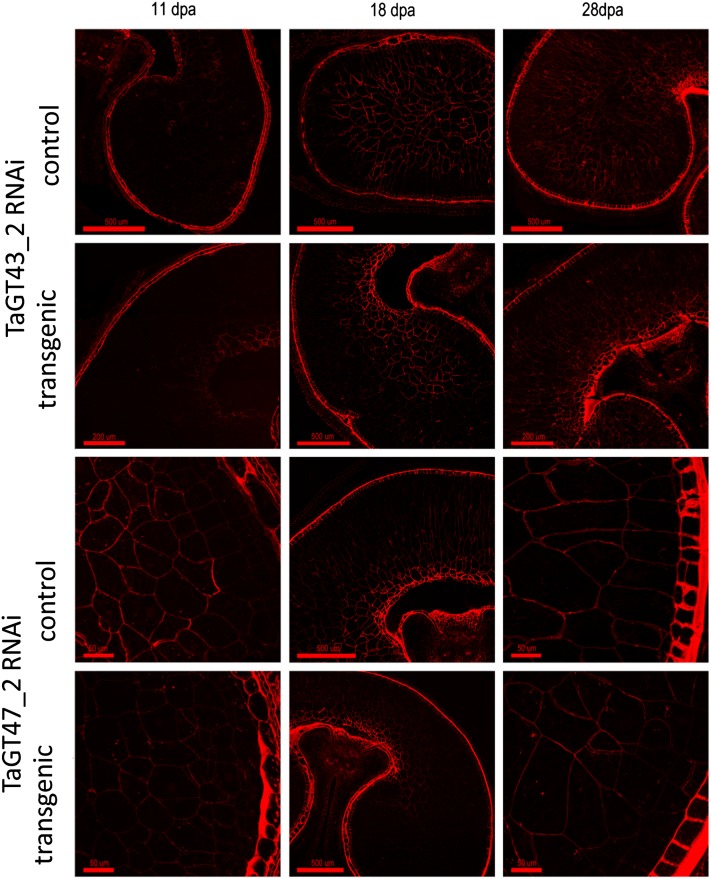
Despite these massive changes in the principal polysaccharide of the endosperm cell wall, the transgenic seed appeared normal and grain weight and germination rates showed no consistent effects (Supplemental Fig. S3). Immunolabeling with the LM11 monoclonal antibody, which labels xylan that has little or no substitution with Ara (McCartney et al., 2005), showed little labeling of the starchy endosperm at 11 DPA, but by 18 DPA, there was substantial labeling in the cell walls of the controls but almost no labeling in large parts of the starchy endosperm in transgenic grain (Fig. 7). In both the TaGT43_2 and TaGT47_2 RNAi samples, labeling tended to be retained in the transfer cell region close to the dorsal crease and lost elsewhere. By 28 DPA, labeling with LM11 appeared to be abolished in the starchy endosperm in both the TaGT43_2 and TaGT47_2 RNAi samples, apart from a small area around the dorsal crease. Immunolabeling with the anti-AX1 monoclonal antibody raised against a mixture of AXOS (Guillon et al., 2004) gave more labeling at 11 DPA but smaller differences between transgenic and control samples (Fig. 8). However, the intensity of labeling was consistently lower in transgenics compared with controls, particularly in the lobe regions of the starchy endosperm; the label also showed apparently thinner cell walls at 28 DPA in transgenics compared with controls (Figs. 7 and 8). Measurements carried out on sections stained with toluidine blue (Supplemental Fig. S4) showed that the cell wall thickness was reduced by about 50% in the transgenic lines (Table II).
Figure 7.
Sections of developing grain from TaGT43_2 RNAi line 6 and TaGT47_2 RNAi line 4 showing comparison of homozygous grain and corresponding azygous controls. Immunolabeling is with the LM11 monoclonal antibody raised against unsubstituted xylan oligosaccharide (McCartney et al., 2005). [See online article for color version of this figure.]
Figure 8.
As for Figure 7, but immunolabeling was with the anti-AX1 monoclonal antibody raised against a mixture of AXOS (Guillon et al., 2004). [See online article for color version of this figure.]
Table II. Cell wall width (in µm) estimated by electronic calipers and a 100× objective on grain sections at 28 DPA stained with toluidine blue (see examples in Supplemental Fig. S4).
Values are averages of multiple observations (n = number of observations) on single grain sections from replicate plants. Lines used were GT43_2 line 6 and GT47_2 line 4.
| Tissue | Construct | Replicate | Control A |
Transgenic H |
H/A | P | ||
|---|---|---|---|---|---|---|---|---|
| Mean ± sd | n | Mean ± sd | n | |||||
| Central endosperm | GT43_2 | A | 2.38 ± 1.22 | 38 | 1.02 ± 0.16 | 24 | 43% | |
| GT43_2 | B | 1.24 ± 0.39 | 13 | |||||
| GT47_2 | A | 2.39 ± 0.43 | 24 | 0.97 ± 0.17 | 25 | 41% | ||
| GT47_2 | B | 2.08 ± 0.44 | 25 | 1.03 ± 0.17 | 25 | 49% | ||
| Average | 2.28 ± 0.45a | 3 | 1.06 ± 0.11a | 4 | 47% | 0.0001 | ||
| Aleurone periclinal | GT43_2 | A | 3.30 ± 0.98 | 12 | ||||
| GT43_2 | B | 2.06 ± 0.28 | 12 | |||||
| GT47_2 | A | 3.45 ± 0.88 | 13 | 2.18 ± 0.60 | 13 | 63% | ||
| GT47_2 | B | 4.09 ± 0.86 | 12 | 1.88 ± 0.49 | 14 | 46% | ||
| Average | 3.61 ± 0.06a | 3 | 2.04 ± 0.16a | 3 | 56% | 0.0035 | ||
| Aleurone anticlinal | GT43_2 | A | 4.57 ± 0.33 | 8 | ||||
| GT43_2 | B | 2.29 ± 0.50 | 12 | |||||
| GT47_2 | A | 4.51 ± 0.74 | 12 | 3.99 ± 0.78 | 13 | 88% | ||
| GT47_2 | B | 4.61 ± 0.86 | 12 | 3.10 ± 0.49 | 14 | 67% | ||
| Average | 4.56 ± 0.08a | 2 | 3.49 ± 0.19a | 4 | 76% | 0.2268 | ||
Mean and sd of means for each grain section shown above.
DISCUSSION
The suppression of TaGT43_2 and TaGT47_2 genes by RNAi resulted in the largest reported decrease of xylan in a grass, and in a cell type where this is the dominant polysaccharide. The effects can be compared with those in Arabidopsis xylan due to mutations in the respective homologs of TaGT43_2 and TaGT47_2, IRX9 and IRX10. Arabidopsis plants carrying these mutations also continue to produce xylan (due to the presence of functionally redundant genes), but in severely decreased amounts and with shorter average chain length (Brown et al., 2007, 2009; Pena et al., 2007; Wu et al., 2010; Lee et al., 2012). However, important differences were identified here: (1) Ara substitution was increased in the transgenic wheat lines, whereas GlcA substitution remains constant in the Arabidopsis mutants; (2) a clear difference in phenotype was identified in the loss of long AX chains in TaGT43_2 but not TaGT47_2 RNAi endosperm, whereas no clear difference between the xylan from irx9 and irx10 Arabidopsis mutants has been reported. These results are discussed in more detail below.
Effect of TaGT43_2 and TaGT47_2 Suppression on Cell Wall Polysaccharides
RNAi suppression of the TaGT43_2 and TaGT47_2 genes decreased the total amount by 50% in some lines, altering its structure such that the Ara-to-Xyl ratio was increased by approximately 25% (Fig. 4, A–C). There was some variation in effects between lines shown by both AXOS abundance and monosaccharide analysis (Fig. 5C); this variation was not explained by the variation between lines of measured effects on transcript abundance (Fig. 2), but this is only a snapshot of abundance for transcripts that vary widely with development.
Allelic variation in the TaGT43_2 and TaGT47_2 genes, or in cis-elements controlling their expression, might be expected to have a large influence on AX and traits dependent on this, such as dietary fiber content and composition and flour extract viscosity. The genes are located on chromosomes 4A, 4B, 4D (TaGT43_2) and 3A, 3B, 3D (TaGT47_2). A consensus quantitative trait locus for AX and viscosity has been mapped to a distal part of the long arm of chromosome 3D (Quraishi et al., 2011). TaGT47_2 D is on the long arm of chromosome 3; greater resolution of its position is not yet possible due to the lack of a physical map for this chromosome, but the ortholog in barley (Hordeum vulgare) MLOC_64806 is localized toward the telomere of 3HL (identified using barley ENSEMBL at http://plants.ensembl.org based on the barley physical map; IBS Consortium, 2012), which suggests that this gene may colocalize with the quantitative trait locus in wheat.
The effects of RNAi suppression on the amounts of oligosaccharides released by xylanase digestion were even more dramatic (Fig. 5A), indicating a decrease in the fraction of AX that was fully digestible to small oligosaccharides by xylanase, as confirmed by MALDI-MS (Fig. 5E). This is explained by the greater Ara substitution in the AX in transgenic lines: cleavage by the GH11 enzyme that was used requires three consecutive unsubstituted Xyl residues, so the more densely substituted AX chains present in the transgenics will have lower frequencies of such cleavage sites.
There was a small increase in 1,3;1,4-β-d-glucan in the transgenic lines that had the greatest decreases in AX (Figs. 4F and 5D). This could be a compensatory effect in response to the depletion of AX in the cell wall; alternatively, it could be due to an accumulation of UDP-Glc in the Golgi lumen caused by the decreased use of substrates for xylan synthesis.
Spatial and Temporal Variations of Effects
Although the HMW1Dx5 promoter drives strong expression at 10 DPA under our standard conditions (Pellny et al., 2012), we have previously observed that the effects of RNAi transgenes driven by this promoter on cell walls do not become evident until 14 DPA (Nemeth et al., 2010); this delay is expected due to the time required for the RNAi machinery to operate and for existing enzymes to turn over. Here, we observed complete abolition of LM11 labeling in most of the starchy endosperm by 18 DPA in the TaGT47_2 line and a similar effect in the most distal layers of the dorsal region and of the lobes in the TaGT43_2 line (Fig. 7). This absence of LM11 labeling is likely due both to the decreased amount of AX and to increased Ara substitution in AX of transgenic lines, since this monoclonal antibody was raised using an unsubstituted penta-1,4-xylanoside glycoprotein (McCartney et al., 2005). This explanation is supported by the labeling pattern of the anti-AX1 monoclonal antibody (which recognizes substituted AXOS; Guillon et al., 2004), as this was never abolished in transgenics, although a decrease in labeling intensity was observable (Fig. 8). LM11 labeling was never abolished in transfer cells and immediately surrounding region cells, which show similar labeling to controls (Fig. 7); this region contains some of the earliest developing cells within the starchy endosperm, and the HMW1Dx5 promoter does not drive expression strongly in transfer cells themselves. In general, the cellular organization and structure of the starchy endosperm of the transgenic lines was remarkably unaffected by the loss of up to 50% of the major cell wall polysaccharide, even though the cell walls were about 50% thinner by 28 DPA (Table II). Although the HMW1Dx5 promoter is specific to the starchy endosperm cells, the aleurone periclinal cell walls were also approximately 40% thinner (Table II); this could be due to the movement of small interfering RNA molecules from the starchy endosperm to the aleurone.
Participation of TaGT43_2 and TaGT47_2 in Xylan Synthetic Machinery
The effects of suppressing TaGT43_2 and TaGT47_2 were similar for most parameters measured (Figs. 4, 5, 7, and 8; Table I). This is in accordance with findings in Arabidopsis, where the effects on xylan of mutations in IRX9, IRX10, and IRX14 (sometimes combined with mutations in close homologs to address functional redundancy) are essentially the same: substantial decreases in xylan content and in xylan chain length. Here, we studied the effects of suppressing homologs of only two of these genes: IRX9 and IRX10. However, the third component, the IRX14 homolog, which we call TaGT43_1, is expressed in wheat endosperm at similar levels to IRX9 (Pellny et al., 2012). The TaGT43_1 protein and another IRX10 homolog were also found in a microsomal complex from wheat seedlings with xylan synthase activity (Zeng et al., 2010). From studies in Arabidopsis, it appears that there is a requirement for all three components to maintain xylan synthase activity (Brown et al., 2007, 2009; Pena et al., 2007; Wu et al., 2010; Lee et al., 2012). Since homologous poplar genes are able to complement for the lack of functional IRX9 or IRX14 (Lee et al., 2011), their function seems to have been conserved in evolution. All the plants that have been fully sequenced have genes for the three components, with the lycophyte S. moellendorffii having the minimal set of three genes (Fig. 1). Our results are consistent with such a xylan synthase complex existing in the starchy endosperm cells of wheat, with components encoded by TaGT43_2 and TaGT47_2 being responsible for the majority of the AX backbone synthesis in this tissue. It seems likely that a third component corresponding to IRX14 would also be essential for normal AX synthesis and that TaGT43_1 probably encodes this component, but this was not tested here.
AX Arabinosylation
The degree of AX substitution was increased as indicated by increases in the AX Ara-to-Xyl ratio of up to 40% (Fig. 4B). This contrasts strongly with irx9 and irx10/irx10-L knockout mutants in Arabidopsis, which maintained the same degree of substitution by methylated or unmethylated GlcA as in the wild-type plants, about 11% of Xyl residues (Brown et al., 2007, 2009), when xylan synthesis was strongly suppressed. It appears that the excess arabinosylation capacity when AX backbone synthesis was suppressed led to regions of highly substituted AX that were not fully digestible by xylanase, leading to even greater decreases in abundance of small AXOS (Fig. 5A). The observation that the unsubstituted X and XX released by xylanase was decreased to about the same degree as total AX, but substituted AXOS was decreased much more (Fig. 5C), may suggest the existence of fractions of AX with low substitution that were not much altered in the transgenics, whereas other fractions with higher Ara substitution tended to become even more highly substituted and, therefore, gave rise to a greater proportion of higher Mr AXOS upon digestion with GH11 (Fig. 5E).
WE-AX Chain Length
Profiles from size-exclusion chromatography suggest that the average chain length of xylan was substantially decreased in the Arabidopsis irx9, irx10, and irx14 mutants (Pena et al., 2007; Brown et al., 2009; Wu et al., 2010). Whereas a rice mutant with inactive OsIRX10 showed no change in the Mr profile of a fraction solubilized by 1 m KOH (Chen et al., 2013), only one of the six IRX10-L rice genes was inactivated, resulting in a 10% decrease in cell wall Xyl, compared with a 60% decrease in the Arabidopsis IRX10/IRX10-L double mutant. Effects on chain length cannot be inferred from profiles of concentration against retention volume alone when substitution is changed, as is the case here, because this will also affect Mr. However, it has been shown that the chain length of wheat grain WE-AX is directly related to [η] for a wide range of Ara-to-Xyl ratios (Dervilly-Pinel et al., 2001). When this relationship was applied, it was clear that both GT43_2 and GT47_2 RNAi transgenic lines did indeed show a substantial decrease in average chain length of WE-AX (Table I; Fig. 6). However, a shoulder in the profiles of [η] for the GT43_2 RNAi transgenic lines, which corresponds to a peak in the control, shows a decrease in the chain length of WE-AX by a factor of more than 2 (Fig. 6, A and B), whereas there is apparently no effect in the GT47_2 RNAi lines (Fig. 6, C and D); similar differences are also seen in the maximum chain length. This suggests that xylan synthase complexes partially deficient in TaGT43_2 lack the capacity to make long AX chains. By contrast, xylan synthase complexes partially deficient in TaGT47_2 can make such long chains, albeit at a much decreased rate. To our knowledge, this is the first finding of a clear difference between the roles of IRX9 and IRX10 homologs, and it is possible that they are only apparent in wheat endosperm WE-AX because of the much greater average chain length of 1,000 to 1,500 Xyl (Saulnier et al., 2007) compared with Arabidopsis stem GX at around 100 Xyl (Pena et al., 2007).
CONCLUSION
RNAi suppression of TaGT43_2 and TaGT47_2 in wheat induces massive changes in the starchy endosperm AX composition. Therefore, variation in the expression of these genes, or the activity of their encoded proteins, will affect important traits such as dietary fiber content and composition and the viscosity of extracts from wheat grain. The effect on AX is consistent with the TaGT43_2 and TaGT47_2 proteins playing key roles in the synthesis of the xylan backbone, but differences in the effects on WE-AX chain length give intriguing clues to the differing roles of these two proteins within the xylan synthesis machinery.
MATERIALS AND METHODS
Plant Growth
Bread wheat (Triticum aestivum ‘Cadenza’) plants were grown in temperature-controlled glasshouse rooms as described (Nemeth et al., 2010).
RNAi Construct Preparation and Transformation
Full-length TaGT43_2 and TaGT47_2 clones were obtained from endosperm complementary DNA as described previously (Anders et al., 2012) but using primers GT43_2F, GT43_2R, GT47_2F, and GT47_2R (Supplemental Table S2). Multiple clones were sequenced as described previously to obtain the consensus sequences shown in Supplemental Figures S1 and S2, which correspond exactly to regions of chromosome-sorted contigs available at www.wheatgenome.org, confirming that the three isoforms originate from homologous chromosomes of the A, B, and D genomes. These sequences have been deposited in the EMBL European Nucleotide Sequence database with the following accession numbers: TaGT43_2D, HF913567; TaGT43_2B, HF913568; TaGT43_2A, HF913569; TaGT47_2B, HF913570; TaGT47_2D, HF913571; TaGT47_2A, HF913572. RNAi constructs with the starchy endosperm-specific HMW1Dx5 promoter were created as described by Nemeth et al. (2010) but using the 446- and 563-bp fragments indicated in Supplemental Figures S1 and S2, respectively, using the PCR primers indicated in Supplemental Table S2. Wheat transformation was carried out by particle bombardment (PDS1000; Bio-Rad) of immature scutella (10–14 DPA) of cv Cadenza according to Sparks and Jones (2009), and zygosity testing was carried out as described previously (Nemeth et al., 2010). For each construct, transgenic lines with segregation consistent with a single insertion locus were identified. In subsequent generations, homozygous and azygous segregants descended from the same original transformant were identified and grown as three replicate pots in block design experiments. Analyses were conducted on T3 or T4 seed from such experiments.
Transcript Analysis
Total RNA was extracted as described (Nemeth et al., 2010). Down-regulation of the transcript was measured as described previously (Anders et al., 2012). In short, developing T4 seeds (T3 for GT47_2 line 6) were harvested at 21 DPA, and RNA from pure endosperm was extracted. In the GT43_2 RNAi lines, the transcript down-regulation was tested using primers prTYW343 and prTYW348, resulting in a 117-bp amplicon. For the GT47_2 transgenics, in addition to the targeted gene (primers prTYW460 and prTYW462; 94-bp fragment), the transcript levels for GT47_1 and GT47_4 were determined, as they also showed some sequence homology to the RNAi fragment used. The primers were prTYW273 and prTYW449 (61 bp) for GT47_1 and prTYW464 and prTYW465 (96 bp) for GT47_4. Three reference genes were used to normalize expression: Ta2526, a stably expressed EST from grain (primers prTYW19 and TYW20), glyceraldehyde-3-phosphate dehydrogenase (primers prTYW270 and prTYW271), and succinate dehydrogenase (primers prTYW209 and prTYW210). All primer sequences are given in Supplemental Table S2.
Cell Wall Analyses
White flour fractions (pure starchy endosperm) were obtained from mature seed as described previously (Anders et al., 2012). Analyses of nonstarch monosaccharide content of white flour samples were conducted following the protocol of Englyst et al. (1994). The methodology for analysis of endosperm AX and 1,3;1,4-β-d-glucan by digestion with endoxylanase and lichenase followed by HPAEC of resultant AXOS was originally developed by Ordaz-Ortiz et al. (2005), and the procedure followed here was as described by Nemeth et al. (2010). The monosaccharide contents of the products of endoxylanase and lichenase digestion (Nemeth et al., 2010; xylanase extractable) were determined as described (Englyst et al., 1994), except that total hydrolysate volume was 7.85 mL. MALDI-MS analyses of AXOS released by digestion were performed on an Autoflex III MALDI-time of flight/time of flight mass spectrometer (Bruker Daltonics) equipped with a Smartbeam laser (355 nm) in positive ion mode with linear detection. Acquisition parameters (laser power, pulsed ion extraction, etc.) were optimized for each sample. Mass spectra were automatically processed with Flex Analysis 3.0 software (Bruker Daltonics). A mixture of 2,5-dihydroxybenzoic acid and N-dimethylaniline was used as ionic matrix, and samples were prepared as described previously (Ropartz et al., 2011).
High-Performance Size-Exclusion Chromatography
For extraction of WE-AX, 500 mg of flour was suspended in 2 mL of water and agitated for 20 min at room temperature; after centrifugation, supernatants were heated for 5 min in a boiling-water bath, filtrated over 0.45 µm, and stabilized to pH 2 with Gly-HCl buffer. Extracts (0.05 mL) were injected on the high-performance size-exclusion chromatography (HPSEC) system. HPSEC was performed at room temperature on a system consisting of a Shodex OH SB-G guard column (Showa Denko) and Shodex OH-Pak SB-805 HQ a columns eluted at 1 mL min−1 with 50 mm sodium nitrate buffer. The Viscotek tri-SEC model 270 was used for light scattering and differential pressure detection, and a Viscotek VE 3580 RI detector was used for the determination of polymer concentration. A refractive index increment per unit concentration increment (dn/dc) value of 0.146 mL g−1 was used for concentration determination. Data were collected with Omnisec 4.5 software (Viscotek), and all calculations on polymer peaks (concentration, [η]) were carried out using Omnisec software. Due to the presence of aggregates in some of the samples, average Mr values from light-scattering measurements were overestimated and, therefore, not used. Conversely, the viscosity detector is not affected by the presence of aggregates; therefore, [η] values were used for comparison of samples.
Microscopy
Transverse slices, approximately 1 mm thick, were cut from developing wheat grains at 11, 18, and 28 DPA and fixed and embedded as described (Pellny et al., 2012). The LM11 antibody was used at a dilution of 1:5; the monoclonal anti-AX1 antibody was used at a 1:25 dilution; secondary antibodies (anti-rat Alexa 633 conjugated and anti-mouse Alexa 568 conjugated [Invitrogen]) were used at a 1:100 dilution. Images were taken using a Zeiss 780 confocal microscope. Cell wall width measurements were done on 1-µm-thick transverse sections stained with toluidine blue prepared from 28-DPA samples. Imaging was done on a Zeiss Axiophot microscope equipped with a QImaging Retiga Exi CCD digital camera using a 100× objective. Measurements of width were done in MetaMorph software (Molecular Devices) using digital calipers.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers HF913567, HF913568, HF913569, HF913570, HF913571, and HF913572.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Alignment of TaGT43_2A, B and D cDNA sequences.
Supplemental Figure S2. Alignment of TaGT47_2A, B and D cDNA sequences.
Supplemental Figure S3. Grain dry weight and germination rates.
Supplemental Figure S4. Example grain sections stained with Toluidine Blue.
Supplemental Table S1. Monosaccharide analyses of endosperm cell walls.
Supplemental Table S2. List of PCR primers.
Acknowledgments
We thank Dr. Stephen Powers (Rothamsted Research) for statistical analyses. We thank D. Ropartz from the Biopolymères, Biologie Structurale platform (INRA Angers-Nantes Center) for mass spectrometry analyses.
Glossary
- Araf
arabinofuranose
- AX
arabinoxylan
- RNAi
RNA interference
- AXOS
arabinoxylan oligosaccharide
- NSP
nonstarch polysaccharides
- WE
water-extractable
- HPAEC
high-performance anion-exchange chromatography
- MALDI
matrix-assisted laser-desorption ionization
- MS
mass spectrometry
- [η]
intrinsic viscosity
- HPSEC
high-performance size-exclusion chromatography
References
- Anders N, Wilkinson MD, Lovegrove A, Freeman J, Tryfona T, Pellny TK, Weimar T, Mortimer JC, Stott K, Baker JM, et al. (2012) Glycosyl transferases in family 61 mediate arabinofuranosyl transfer onto xylan in grasses. Proc Natl Acad Sci USA 109: 989–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DM, Goubet F, Wong VW, Goodacre R, Stephens E, Dupree P, Turner SR. (2007) Comparison of five xylan synthesis mutants reveals new insight into the mechanisms of xylan synthesis. Plant J 52: 1154–1168 [DOI] [PubMed] [Google Scholar]
- Brown DM, Zhang ZN, Stephens E, Dupree P, Turner SR. (2009) Characterization of IRX10 and IRX10-like reveals an essential role in glucuronoxylan biosynthesis in Arabidopsis. Plant J 57: 732–746 [DOI] [PubMed] [Google Scholar]
- Carpita NC. (1996) Structure and biogenesis of the cell walls of grasses. Annu Rev Plant Physiol Plant Mol Biol 47: 445–476 [DOI] [PubMed] [Google Scholar]
- Chen X, Vega-Sánchez ME, Verhertbruggen Y, Chiniquy D, Canlas PE, Fagerström A, Prak L, Christensen U, Oikawa A, Chern M, et al. (2013) Inactivation of OsIRX10 leads to decreased xylan content in ricestem cell walls and improved biomass saccharification. Mol Plant 6: 570–573 [DOI] [PubMed] [Google Scholar]
- Chiniquy D, Varanasi P, Oh T, Harholt J, Katnelson J, Singh S, Auer M, Simmons B, Adams PD, Scheller HV, et al. (2013) Three novel rice genes closely related to the Arabidopsis IRX9, IRX9L, and IRX14 genes and their roles in xylan biosynthesis. Front Plant Sci 4: 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dervilly-Pinel G, Thibault JF, Saulnier L. (2001) Experimental evidence for a semi-flexible conformation for arabinoxylans. Carbohydr Res 330: 365–372 [DOI] [PubMed] [Google Scholar]
- Ebringerova A, Hromadkova Z, Heinze T. (2005) Hemicellulose. Adv Polym Sci 186: 1–67 [Google Scholar]
- Englyst HN, Quigley ME, Hudson GJ. (1994) Determination of dietary fiber as nonstarch polysaccharides with gas-liquid-chromatographic, high-performance liquid-chromatographic or spectrophotometric measurement of constituent sugars. Analyst (Lond) 119: 1497–1509 [DOI] [PubMed] [Google Scholar]
- Faure R, Courtin CM, Delcour JA, Dumon C, Faulds CB, Fincher GB, Fort S, Fry SC, Halila S, Kabel MA, et al. (2009) A brief and informationally rich naming system for oligosaccharide motifs of heteroxylans found in plant cell walls. Aust J Chem 62: 533–537 [Google Scholar]
- Goodstein DM, Shu SQ, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, et al. (2012) Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res 40: D1178–D1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillon F, Tranquet O, Quillien L, Utille JP, Ortiz JJO, Saulnier L. (2004) Generation of polyclonal and monoclonal antibodies against arabinoxylans and their use for immunocytochemical location of arabinoxylans in cell walls of endosperm of wheat. J Cereal Sci 40: 167–182 [Google Scholar]
- IBS Consortium (2012) A physical, genetic and functional sequence assembly of the barley genome. Nature 491: 711–716 [DOI] [PubMed] [Google Scholar]
- Jensen JK, Kim H, Cocuron JC, Orler R, Ralph J, Wilkerson CG. (2011) The DUF579 domain containing proteins IRX15 and IRX15-L affect xylan synthesis in Arabidopsis. Plant J 66: 387–400 [DOI] [PubMed] [Google Scholar]
- Konishi T, Takeda T, Miyazaki Y, Ohnishi-Kameyama M, Hayashi T, O’Neill MA, Ishii T. (2007) A plant mutase that interconverts UDP-arabinofuranose and UDP-arabinopyranose. Glycobiology 17: 345–354 [DOI] [PubMed] [Google Scholar]
- Lee C, Zhong RQ, Ye ZH. (2012) Arabidopsis family GT43 members are xylan xylosyltransferases required for the elongation of the xylan backbone. Plant Cell Physiol 53: 135–143 [DOI] [PubMed] [Google Scholar]
- Lee CH, Teng QC, Zhong RQ, Ye ZH. (2011) Molecular dissection of xylan biosynthesis during wood formation in poplar. Mol Plant 4: 730–747 [DOI] [PubMed] [Google Scholar]
- McCartney L, Marcus SE, Knox JP. (2005) Monoclonal antibodies to plant cell wall xylans and arabinoxylans. J Histochem Cytochem 53: 543–546 [DOI] [PubMed] [Google Scholar]
- Mortimer JC, Miles GP, Brown DM, Zhang ZN, Segura MP, Weimar T, Yu XL, Seffen KA, Stephens E, Turner SR, et al. (2010) Absence of branches from xylan in Arabidopsis gux mutants reveals potential for simplification of lignocellulosic biomass. Proc Natl Acad Sci USA 107: 17409–17414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemeth C, Freeman J, Jones HD, Sparks C, Pellny TK, Wilkinson MD, Dunwell J, Andersson AAM, Aman P, Guillon F, et al. (2010) Down-regulation of the CSLF6 gene results in decreased (1,3;1,4)-β-D-glucan in endosperm of wheat. Plant Physiol 152: 1209–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ordaz-Ortiz JJ, Devaux MF, Saulnier L. (2005) Classification of wheat varieties based on structural features of arabinoxylans as revealed by endoxylanase treatment of flour and grain. J Agric Food Chem 53: 8349–8356 [DOI] [PubMed] [Google Scholar]
- Ordaz-Ortiz JJ, Saulnier L. (2005) Structural variability of arabinoxylans from wheat flour: comparison of water-extractable and xylanase-extractable arabinoxylans. J Cereal Sci 42: 119–125 [Google Scholar]
- Pellny TK, Lovegrove A, Freeman J, Tosi P, Love CG, Knox JP, Shewry PR, Mitchell RAC. (2012) Cell walls of developing wheat starchy endosperm: comparison of composition and RNA-Seq transcriptome. Plant Physiol 158: 612–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peña MJ, Zhong RQ, Zhou GK, Richardson EA, O’Neill MA, Darvill AG, York WS, Ye ZH. (2007) Arabidopsis irregular xylem8 and irregular xylem9: implications for the complexity of glucuronoxylan biosynthesis. Plant Cell 19: 549–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quraishi UM, Murat F, Abrouk M, Pont C, Confolent C, Oury FX, Ward J, Boros D, Gebruers K, Delcour JA, et al. (2011) Combined meta-genomics analyses unravel candidate genes for the grain dietary fiber content in bread wheat (Triticum aestivum L.). Funct Integr Genomics 11: 71–83 [DOI] [PubMed] [Google Scholar]
- Ropartz D, Bodet PE, Przybylski C, Gonnet F, Daniel R, Fer M, Helbert W, Bertrand D, Rogniaux H. (2011) Performance evaluation on a wide set of matrix-assisted laser desorption ionization matrices for the detection of oligosaccharides in a high-throughput mass spectrometric screening of carbohydrate depolymerizing enzymes. Rapid Commun Mass Spectrom 25: 2059–2070 [DOI] [PubMed] [Google Scholar]
- Saulnier L, Sado PE, Branlard G, Charmet G, Guillon F. (2007) Wheat arabinoxylans: Exploiting variation in amount and composition to develop enhanced varieties. J Cereal Sci 46: 261–281 [Google Scholar]
- Scheller HV, Ulvskov P. (2010) Hemicelluloses. Annu Rev Plant Biol 61: 263–289 [DOI] [PubMed] [Google Scholar]
- Sparks CA, Jones HD (2009) Biolistics transformation of wheat. In HD Jones, PR Shewry, eds, Transgenic Wheat, Barley and Oats. Humana Press, New York, NY, pp 71–92 [Google Scholar]
- Topping D. (2007) Cereal complex carbohydrates and their contribution to human health. J Cereal Sci 46: 220–229 [Google Scholar]
- Wu A-M, Rihouey C, Seveno M, Hörnblad E, Singh SK, Matsunaga T, Ishii T, Lerouge P, Marchant A. (2009) The Arabidopsis IRX10 and IRX10-LIKE glycosyltransferases are critical for glucuronoxylan biosynthesis during secondary cell wall formation. Plant J 57: 718–731 [DOI] [PubMed] [Google Scholar]
- Wu AM, Hörnblad E, Voxeur A, Gerber L, Rihouey C, Lerouge P, Marchant A. (2010) Analysis of the Arabidopsis IRX9/IRX9-L and IRX14/IRX14-L pairs of glycosyltransferase genes reveals critical contributions to biosynthesis of the hemicellulose glucuronoxylan. Plant Physiol 153: 542–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- York WS, O’Neill MA. (2008) Biochemical control of xylan biosynthesis: which end is up? Curr Opin Plant Biol 11: 258–265 [DOI] [PubMed] [Google Scholar]
- Zeng W, Jiang N, Nadella R, Killen TL, Nadella V, Faik A. (2010) A glucurono(arabino)xylan synthase complex from wheat contains members of the GT43, GT47, and GT75 families and functions cooperatively. Plant Physiol 154: 78–97 [DOI] [PMC free article] [PubMed] [Google Scholar]