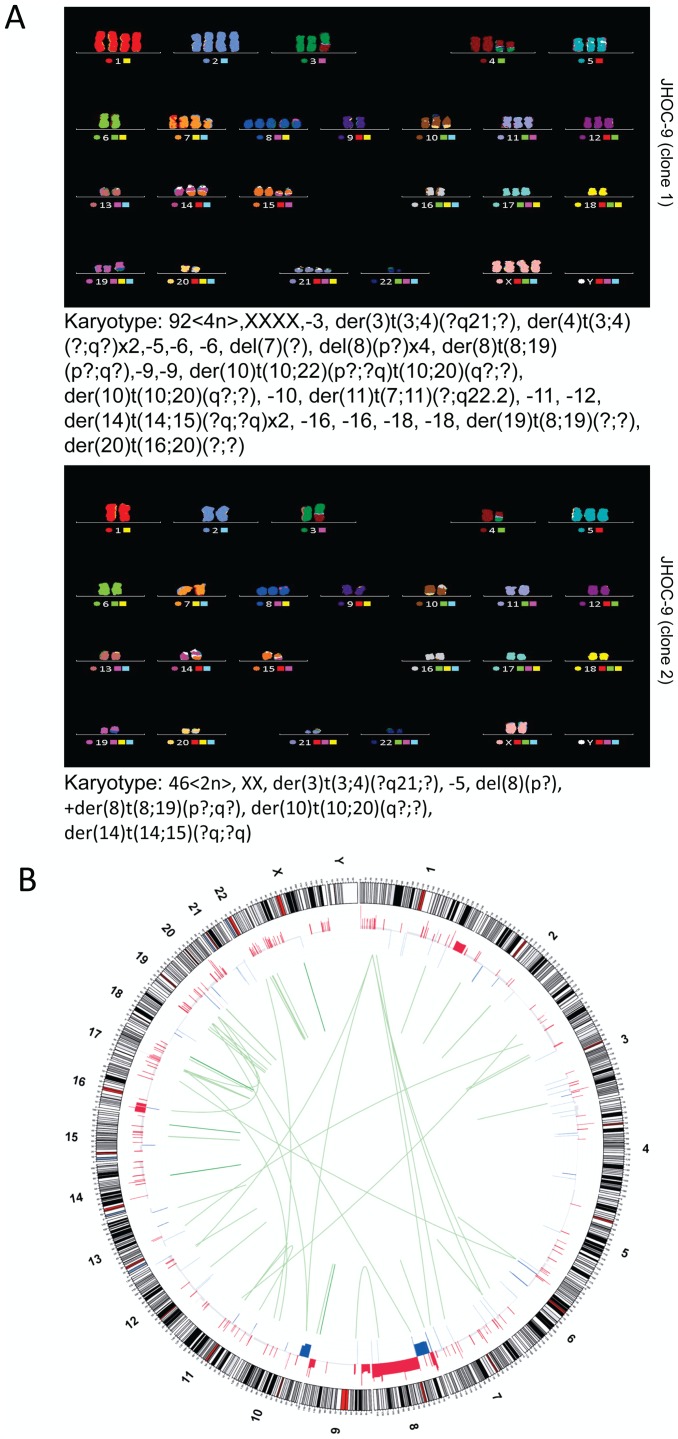
Figure 3. Genomic structure of CCC cell line JHOC-9. (A) 24 color FISH analysis suggested the presence of two dominant clones; one near-diploid and one near-tetraploid in the JHOC-9 CCC cell line.
A number of translocations and rearrangements can be seen in each representative clone. The complex karyotype of each dominant clone is noted below the corresponding 24-colour FISH results. Not all derivative chromosomes were identifiable resulting in a large number of ambiguous translocations and fragments (denoted by question marks in the karyotype notations). (B) Circos plot of RNAseq data and deFuse analysis depicting expressed genomic rearrangements in the JHOC-9 cell line. Translocations seen in the 24-color FISH profile are also visible as expressed transcripts including t(8;19) observed in both 2N and 4N dominant clones. No recurrent translocations were seen across our series (see also Table S3).