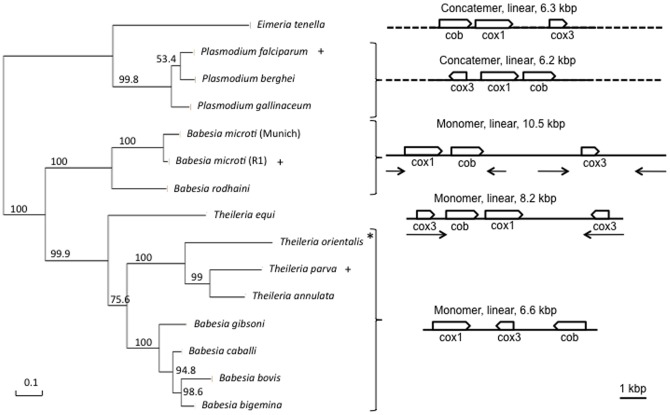
Figure 8. Phylogenetic analysis on the left panel is based on a combination of three protein sequences encoded by the genome of different apicomplexan mitochondria (cox1+cob+cox3).
About 63% of the sites were conserved for the phylogenetic analysis. The scale indicates the inferred number of substitutions. Boostrap values are associated to each branch. The unrooted tree was calculated using the Phygeny.fr web site and default options of the one-click procedure. The schematic structures of mitochondrial genomes in apicomplexa (right panel) are based on species marked by a “+” in (A). Inverted repeats are represented by arrows. T. parva is presenting short inveted repeats that are not described in other Theileria and true-Babesia species. Dashed lines figure out the concatenated form of the molecule. * T. orientalis is presenting an inversion of the cox3 gene. The gene order is highly conserved among Plasmodium species [47]. Variations are associated to rRNA genes and resolution site. The P. falciparum genome organization is also present in other species such as P. floridensis, P. mexicanum or P. reichenovi. A slight variation in the molecule structure of P. berghei and P. gallineum does not change the gene order (Fig. S3 in Supporting Information S1). This later organization is present in other species such as P. fragile, P. knwolesi, P. sinium, P. vivax or P. yoelii.