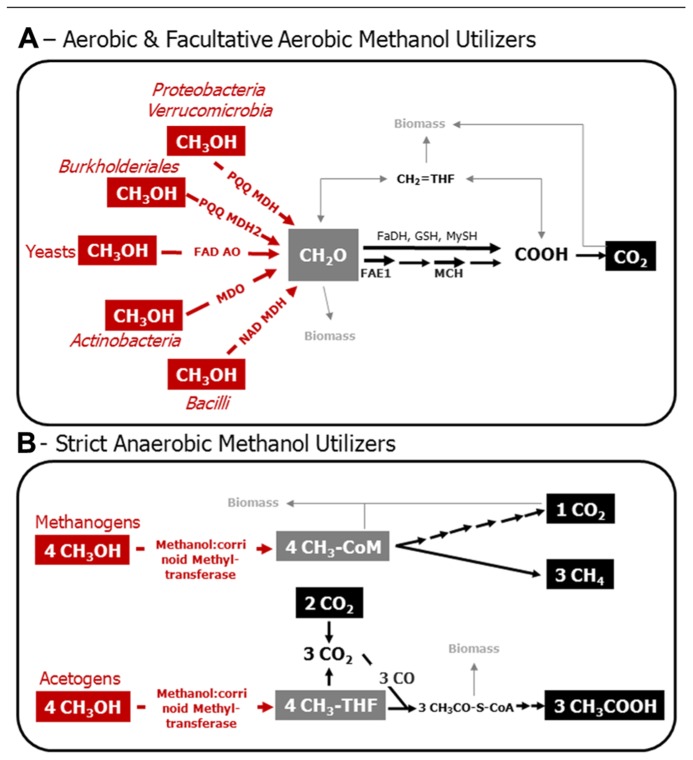
FIGURE 1.
Model of the C1 metabolism of aerobic and facultatively aerobic methanol utilizers (A), and of methanol-utilizing methanogens and acetogens (B). Various Bacteria that employ PQQ MDH and PQQ MDH2 utilize nitrate besides molecular oxygen, and are, thus, facultative aerobes. Red, enzymatic reactions indicative for methanol utilization. Gray, metabolic crossing points to anabolic pathways. Structural genes are targets for the molecular assessment of methanol-utilizing microorganisms in the environment. PQQ MDH, PQQ-dependent methanol dehydrogenase; PQQ MDH2, alternative PQQ-dependent methanol dehydrogenase in Methylibium andBurkholderia strains; FAD AO, FAD-dependent alcohol oxidoreductase; MDO, methanol oxidoreductase; NAD MDH, NAD-dependent methanol dehydrogenase; FAE1, tetrahydromethanopterin-dependent formaldehyde activating enzyme; MCH, tetrahydromethanopterin-dependent methenyl-methylene cyclohydrolase; FaDH, formaldehyde dehydrogenase; GSH, glutathione-dependent formaldehyde oxidation; MySH, mycothiol-dependent oxidation of formaldehyde in yeast; CH2 = HF4, methylene tetrahydrofolate; CH3-THF, methyl-tetrahydrofolate; CH3-CoM, methyl-coenzyme M; CH3OH, methanol; CH2O, formaldehyde; COOH formate; CO2, carbon dioxide; CO, carbon monoxide; CH3CO–S–CoA, acetyl coenzyme A; CH3COOH, acetic acid. Biomass, assimilation of carbon occurs at the level of formaldehyde and/or on carbon dioxide in aerobic methylotrophs; pathways involved are the Calvin–Benson–Bassham cycle, ribulose-monophosphate pathway, and the serine cycle. Assimilation of carbon in methanogens is mediated by a unique reductive acetyl-CoA-pathway, whereas acetogens form acetyl-CoA as an intermediate that can be used for biosnthesis. This figure is based on previous articles (Thauer, 1998; Ding et al., 2002; Hagemeier et al., 2006; Das et al., 2007; Drake et al., 2008; Chistoserdova et al., 2009; Chistoserdova, 2011; Gvozdev et al., 2012).