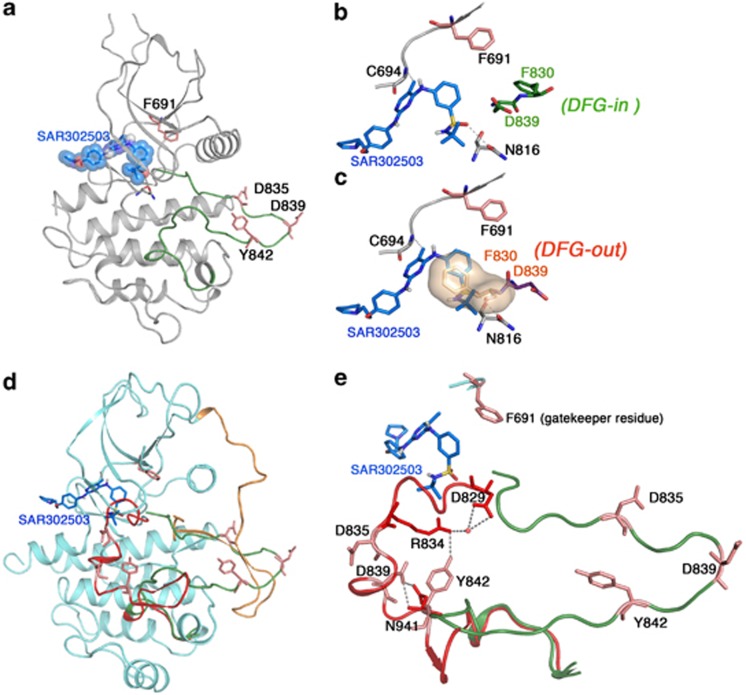
Figure 2.
SAR302503 binds to ATP site, an active conformation of the FLT3 kinase. (a) Ribbon depiction of a structural model of FLT3-SAR302503 using the coordinates of the inactive FLT3 kinase (PDB:3CS9), ABL (PDB:2Z60) and JAK2 (PDB:2B7A). Activation loop is shown as green and extended conformation during active state. (b) A close-up view of the active site of the FLT3 kinase showing the interaction of SAR302503 by hydrogen bonding with Cys 694 and Asn 816. Further, SAR302503 can bind only in DFG-in conformational state (residues shown in green sticks), in which phenylalanine of the DFG motif is aligned in such a way that it coordinates with hydrophobic spine residues to stabilize the active state, a common feature in all kinases suggesting that SAR302503 can only bind to active state and thus can be classified as a type I inhibitor. (c) Active site showing the steric clash of the benzene sulfonamide group of SAR302503 with Phe 830 in DGF-out conformation (F830 from the DFG motif shown in surface). (d) Structure of autoinhibited FLT3 shown in ribbon with modeled activation loops in active conformation (shown in green) and in inactive conformation (shown in red). The juxtamembrane domain (orange) nearly spans the length of the kinase molecule and seemingly displaces the activation loop from the active state to favor inactive conformation. Amino-acid residues that were mutated to confer drug resistance are shown as sticks. (e) Conformation of activation loops in active (green) and inactive (red) states showing interaction of Y842 with D829 (DFG motif) through R834 and D839 with N934 to stabilize the inactive conformation, and thus mutation at these sites may favor or stabilize active state that will be more sensitive to inhibition by type I inhibitors. In support of this model, variant Y842H showing increased autophosphorylation in untreated sample, whereas D839G did not show significant change suggesting that it only destabilizes the inactive conformation and may be unable to stabilize the fully active conformation as we have observed in the Y842H. Nonetheless, it supports that mutant D839G stabilizes a conformation favoring efficient SAR302503 binding that reflects in hypersensitive response. The hypersensitive response of variant D839G is less than Y842H suggesting that the later is stabilizing a fully active state that binds the drug more efficiently. Nevertheless, these speculations require detail structural studies for better understanding and developing next-generation FLT3 inhibitors.